
Wenzhouxiangella marina
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Wenzhouxiangellaceae; Wenzhouxiangella
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
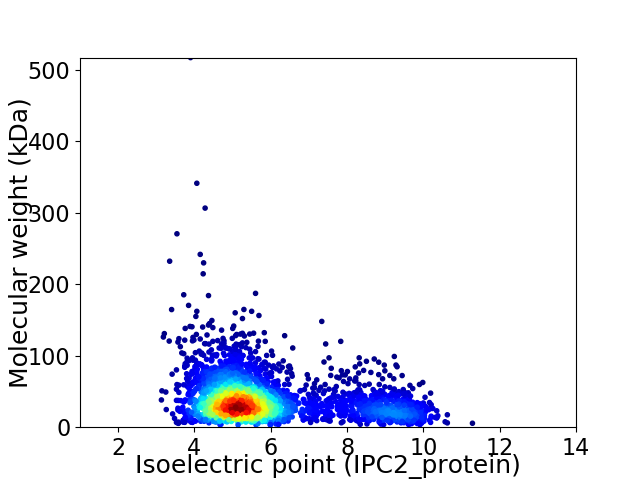
Virtual 2D-PAGE plot for 3023 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K0XUA3|A0A0K0XUA3_9GAMM Uncharacterized protein OS=Wenzhouxiangella marina OX=1579979 GN=WM2015_913 PE=4 SV=1
MM1 pKa = 7.68AAGLLATHH9 pKa = 7.65AIATVEE15 pKa = 4.02YY16 pKa = 9.61DD17 pKa = 3.35QDD19 pKa = 3.78VTPTIIFGAGNNNGAFTTDD38 pKa = 2.7RR39 pKa = 11.84FNGVEE44 pKa = 3.35IGLRR48 pKa = 11.84AKK50 pKa = 10.39LRR52 pKa = 11.84YY53 pKa = 9.55DD54 pKa = 3.64EE55 pKa = 5.61NGVPQNVFNSNGDD68 pKa = 3.22GSYY71 pKa = 9.42TFYY74 pKa = 10.85TRR76 pKa = 11.84SRR78 pKa = 11.84PNFPDD83 pKa = 3.58GQRR86 pKa = 11.84GEE88 pKa = 4.52WSFEE92 pKa = 3.7WAVNTDD98 pKa = 3.9FDD100 pKa = 5.05GSSLLNLDD108 pKa = 4.04DD109 pKa = 5.14LSYY112 pKa = 11.3EE113 pKa = 4.4LGLDD117 pKa = 3.7ADD119 pKa = 4.99PGPGTNFLVFDD130 pKa = 6.08PITPSAEE137 pKa = 3.99APCWDD142 pKa = 3.89HH143 pKa = 7.57AIGNNGTGNGGGTAVNCITDD163 pKa = 4.0PNPGQSYY170 pKa = 11.16AGLLTDD176 pKa = 5.04NNVAQNSWQYY186 pKa = 11.48GFFLIDD192 pKa = 3.38EE193 pKa = 4.72LADD196 pKa = 3.59YY197 pKa = 10.7DD198 pKa = 3.67PRR200 pKa = 11.84LPGRR204 pKa = 11.84YY205 pKa = 7.58TIYY208 pKa = 10.58LQASNGSGVVARR220 pKa = 11.84SEE222 pKa = 4.0IEE224 pKa = 4.36VIVVDD229 pKa = 4.49PPLAFDD235 pKa = 3.92QNVTPEE241 pKa = 4.66AIFGSGNANGEE252 pKa = 4.1FTTDD256 pKa = 2.85RR257 pKa = 11.84RR258 pKa = 11.84NDD260 pKa = 3.05IEE262 pKa = 4.86LGLRR266 pKa = 11.84GKK268 pKa = 10.19LRR270 pKa = 11.84YY271 pKa = 8.88NASGVPEE278 pKa = 3.9NTFNSNGDD286 pKa = 3.36GTYY289 pKa = 10.7SFAPRR294 pKa = 11.84LVPGGSPLRR303 pKa = 11.84AEE305 pKa = 4.68WGFEE309 pKa = 3.43WSVNTNYY316 pKa = 10.23QGGAGPNVDD325 pKa = 3.27AYY327 pKa = 9.76TYY329 pKa = 10.93EE330 pKa = 4.91LGLDD334 pKa = 3.89ADD336 pKa = 4.56PSLGTDD342 pKa = 4.07FLVFDD347 pKa = 6.28PITPTAEE354 pKa = 4.22VPCWDD359 pKa = 3.72HH360 pKa = 8.34AMGNNGTPNGGGTSANCLGDD380 pKa = 3.84PNAGATYY387 pKa = 10.81QNLIANNNVAQNSWRR402 pKa = 11.84YY403 pKa = 9.05EE404 pKa = 3.94FFASGPIAGFDD415 pKa = 3.53PTVDD419 pKa = 3.11GSYY422 pKa = 10.43RR423 pKa = 11.84IYY425 pKa = 10.67LAAYY429 pKa = 8.13NADD432 pKa = 3.33GVEE435 pKa = 4.37VARR438 pKa = 11.84SDD440 pKa = 3.03IDD442 pKa = 3.45ILVGSASGGTVSDD455 pKa = 4.52VEE457 pKa = 4.82LSMSTTATGTQFTGDD472 pKa = 4.01AITYY476 pKa = 9.22QLVASNTDD484 pKa = 3.28LAKK487 pKa = 10.47AANVSILNVLPDD499 pKa = 3.7NLSFVAGSCDD509 pKa = 3.65DD510 pKa = 3.82GSVANVAGQSVDD522 pKa = 3.72FALADD527 pKa = 4.03LPSGASTICTIDD539 pKa = 3.39TVVASSGTIVNSASVAADD557 pKa = 3.26NDD559 pKa = 3.3GDD561 pKa = 4.36ANNNAASVRR570 pKa = 11.84LLGVIEE576 pKa = 4.45SVALTGDD583 pKa = 3.31IPSPTDD589 pKa = 3.16NDD591 pKa = 3.36YY592 pKa = 10.87TRR594 pKa = 11.84INDD597 pKa = 3.47VVQIAGPGDD606 pKa = 3.66QIVLNGVFDD615 pKa = 4.3WNEE618 pKa = 3.98SNAFASWALGSDD630 pKa = 4.55GIDD633 pKa = 3.22GTVDD637 pKa = 2.63DD638 pKa = 4.12WTIYY642 pKa = 10.74VPDD645 pKa = 4.32GLADD649 pKa = 3.68LTITATAPGDD659 pKa = 3.75ATIKK663 pKa = 10.95GPGDD667 pKa = 3.35LAGVDD672 pKa = 3.89LEE674 pKa = 4.89GFLLVYY680 pKa = 8.25GTNPGLEE687 pKa = 3.79ISNLVIEE694 pKa = 5.26DD695 pKa = 3.49FDD697 pKa = 4.06VAIGIYY703 pKa = 10.43YY704 pKa = 10.39NGGGVNVYY712 pKa = 11.16DD713 pKa = 4.19NLTIIDD719 pKa = 4.35NFIAMPRR726 pKa = 11.84DD727 pKa = 3.48VAGNSQGGEE736 pKa = 3.71AFQNIGIHH744 pKa = 6.14YY745 pKa = 10.13SFGDD749 pKa = 3.56NILIARR755 pKa = 11.84NVIEE759 pKa = 4.57IPGDD763 pKa = 3.82SASTASLRR771 pKa = 11.84AAQVAMQSNTSGGAYY786 pKa = 9.07EE787 pKa = 4.3GLVIEE792 pKa = 5.02DD793 pKa = 3.44NEE795 pKa = 4.08IRR797 pKa = 11.84ILLAQAEE804 pKa = 4.53IPAGIIGIWEE814 pKa = 4.2NGNAHH819 pKa = 6.39TSNITVRR826 pKa = 11.84NNRR829 pKa = 11.84FINLDD834 pKa = 3.61PANDD838 pKa = 3.81PSLNDD843 pKa = 3.17QEE845 pKa = 4.69AFRR848 pKa = 11.84ITSHH852 pKa = 6.45SSASSTTRR860 pKa = 11.84YY861 pKa = 8.54EE862 pKa = 3.8GNYY865 pKa = 10.47AEE867 pKa = 4.81GANVGYY873 pKa = 10.36GWLSFDD879 pKa = 3.75TYY881 pKa = 11.43GADD884 pKa = 3.51FSTRR888 pKa = 11.84DD889 pKa = 3.62PIEE892 pKa = 4.25FVSNTAVDD900 pKa = 3.85NLTGILIDD908 pKa = 3.95SNGAADD914 pKa = 4.32LSCNRR919 pKa = 11.84IHH921 pKa = 7.49GNDD924 pKa = 3.87LGLSNITQAGRR935 pKa = 11.84ISLADD940 pKa = 4.22DD941 pKa = 3.37NWWGCNAGPNAGDD954 pKa = 3.67CDD956 pKa = 4.34AYY958 pKa = 10.57DD959 pKa = 3.57TGLTTDD965 pKa = 2.91RR966 pKa = 11.84WLVAGLTADD975 pKa = 3.78AGTVLINSTTGLNLDD990 pKa = 4.25LRR992 pKa = 11.84SNSDD996 pKa = 3.21GNEE999 pKa = 4.04VTSCTLPATPVILAANEE1016 pKa = 4.34GSVTPAVGATSAALLDD1032 pKa = 4.1ADD1034 pKa = 3.85YY1035 pKa = 8.08TAPGFATGDD1044 pKa = 3.79TVTVTVDD1051 pKa = 3.47AEE1053 pKa = 4.31VLTVDD1058 pKa = 4.44FTVEE1062 pKa = 4.18LPVDD1066 pKa = 4.06SIFNDD1071 pKa = 3.35RR1072 pKa = 11.84FEE1074 pKa = 4.09NN1075 pKa = 3.72
MM1 pKa = 7.68AAGLLATHH9 pKa = 7.65AIATVEE15 pKa = 4.02YY16 pKa = 9.61DD17 pKa = 3.35QDD19 pKa = 3.78VTPTIIFGAGNNNGAFTTDD38 pKa = 2.7RR39 pKa = 11.84FNGVEE44 pKa = 3.35IGLRR48 pKa = 11.84AKK50 pKa = 10.39LRR52 pKa = 11.84YY53 pKa = 9.55DD54 pKa = 3.64EE55 pKa = 5.61NGVPQNVFNSNGDD68 pKa = 3.22GSYY71 pKa = 9.42TFYY74 pKa = 10.85TRR76 pKa = 11.84SRR78 pKa = 11.84PNFPDD83 pKa = 3.58GQRR86 pKa = 11.84GEE88 pKa = 4.52WSFEE92 pKa = 3.7WAVNTDD98 pKa = 3.9FDD100 pKa = 5.05GSSLLNLDD108 pKa = 4.04DD109 pKa = 5.14LSYY112 pKa = 11.3EE113 pKa = 4.4LGLDD117 pKa = 3.7ADD119 pKa = 4.99PGPGTNFLVFDD130 pKa = 6.08PITPSAEE137 pKa = 3.99APCWDD142 pKa = 3.89HH143 pKa = 7.57AIGNNGTGNGGGTAVNCITDD163 pKa = 4.0PNPGQSYY170 pKa = 11.16AGLLTDD176 pKa = 5.04NNVAQNSWQYY186 pKa = 11.48GFFLIDD192 pKa = 3.38EE193 pKa = 4.72LADD196 pKa = 3.59YY197 pKa = 10.7DD198 pKa = 3.67PRR200 pKa = 11.84LPGRR204 pKa = 11.84YY205 pKa = 7.58TIYY208 pKa = 10.58LQASNGSGVVARR220 pKa = 11.84SEE222 pKa = 4.0IEE224 pKa = 4.36VIVVDD229 pKa = 4.49PPLAFDD235 pKa = 3.92QNVTPEE241 pKa = 4.66AIFGSGNANGEE252 pKa = 4.1FTTDD256 pKa = 2.85RR257 pKa = 11.84RR258 pKa = 11.84NDD260 pKa = 3.05IEE262 pKa = 4.86LGLRR266 pKa = 11.84GKK268 pKa = 10.19LRR270 pKa = 11.84YY271 pKa = 8.88NASGVPEE278 pKa = 3.9NTFNSNGDD286 pKa = 3.36GTYY289 pKa = 10.7SFAPRR294 pKa = 11.84LVPGGSPLRR303 pKa = 11.84AEE305 pKa = 4.68WGFEE309 pKa = 3.43WSVNTNYY316 pKa = 10.23QGGAGPNVDD325 pKa = 3.27AYY327 pKa = 9.76TYY329 pKa = 10.93EE330 pKa = 4.91LGLDD334 pKa = 3.89ADD336 pKa = 4.56PSLGTDD342 pKa = 4.07FLVFDD347 pKa = 6.28PITPTAEE354 pKa = 4.22VPCWDD359 pKa = 3.72HH360 pKa = 8.34AMGNNGTPNGGGTSANCLGDD380 pKa = 3.84PNAGATYY387 pKa = 10.81QNLIANNNVAQNSWRR402 pKa = 11.84YY403 pKa = 9.05EE404 pKa = 3.94FFASGPIAGFDD415 pKa = 3.53PTVDD419 pKa = 3.11GSYY422 pKa = 10.43RR423 pKa = 11.84IYY425 pKa = 10.67LAAYY429 pKa = 8.13NADD432 pKa = 3.33GVEE435 pKa = 4.37VARR438 pKa = 11.84SDD440 pKa = 3.03IDD442 pKa = 3.45ILVGSASGGTVSDD455 pKa = 4.52VEE457 pKa = 4.82LSMSTTATGTQFTGDD472 pKa = 4.01AITYY476 pKa = 9.22QLVASNTDD484 pKa = 3.28LAKK487 pKa = 10.47AANVSILNVLPDD499 pKa = 3.7NLSFVAGSCDD509 pKa = 3.65DD510 pKa = 3.82GSVANVAGQSVDD522 pKa = 3.72FALADD527 pKa = 4.03LPSGASTICTIDD539 pKa = 3.39TVVASSGTIVNSASVAADD557 pKa = 3.26NDD559 pKa = 3.3GDD561 pKa = 4.36ANNNAASVRR570 pKa = 11.84LLGVIEE576 pKa = 4.45SVALTGDD583 pKa = 3.31IPSPTDD589 pKa = 3.16NDD591 pKa = 3.36YY592 pKa = 10.87TRR594 pKa = 11.84INDD597 pKa = 3.47VVQIAGPGDD606 pKa = 3.66QIVLNGVFDD615 pKa = 4.3WNEE618 pKa = 3.98SNAFASWALGSDD630 pKa = 4.55GIDD633 pKa = 3.22GTVDD637 pKa = 2.63DD638 pKa = 4.12WTIYY642 pKa = 10.74VPDD645 pKa = 4.32GLADD649 pKa = 3.68LTITATAPGDD659 pKa = 3.75ATIKK663 pKa = 10.95GPGDD667 pKa = 3.35LAGVDD672 pKa = 3.89LEE674 pKa = 4.89GFLLVYY680 pKa = 8.25GTNPGLEE687 pKa = 3.79ISNLVIEE694 pKa = 5.26DD695 pKa = 3.49FDD697 pKa = 4.06VAIGIYY703 pKa = 10.43YY704 pKa = 10.39NGGGVNVYY712 pKa = 11.16DD713 pKa = 4.19NLTIIDD719 pKa = 4.35NFIAMPRR726 pKa = 11.84DD727 pKa = 3.48VAGNSQGGEE736 pKa = 3.71AFQNIGIHH744 pKa = 6.14YY745 pKa = 10.13SFGDD749 pKa = 3.56NILIARR755 pKa = 11.84NVIEE759 pKa = 4.57IPGDD763 pKa = 3.82SASTASLRR771 pKa = 11.84AAQVAMQSNTSGGAYY786 pKa = 9.07EE787 pKa = 4.3GLVIEE792 pKa = 5.02DD793 pKa = 3.44NEE795 pKa = 4.08IRR797 pKa = 11.84ILLAQAEE804 pKa = 4.53IPAGIIGIWEE814 pKa = 4.2NGNAHH819 pKa = 6.39TSNITVRR826 pKa = 11.84NNRR829 pKa = 11.84FINLDD834 pKa = 3.61PANDD838 pKa = 3.81PSLNDD843 pKa = 3.17QEE845 pKa = 4.69AFRR848 pKa = 11.84ITSHH852 pKa = 6.45SSASSTTRR860 pKa = 11.84YY861 pKa = 8.54EE862 pKa = 3.8GNYY865 pKa = 10.47AEE867 pKa = 4.81GANVGYY873 pKa = 10.36GWLSFDD879 pKa = 3.75TYY881 pKa = 11.43GADD884 pKa = 3.51FSTRR888 pKa = 11.84DD889 pKa = 3.62PIEE892 pKa = 4.25FVSNTAVDD900 pKa = 3.85NLTGILIDD908 pKa = 3.95SNGAADD914 pKa = 4.32LSCNRR919 pKa = 11.84IHH921 pKa = 7.49GNDD924 pKa = 3.87LGLSNITQAGRR935 pKa = 11.84ISLADD940 pKa = 4.22DD941 pKa = 3.37NWWGCNAGPNAGDD954 pKa = 3.67CDD956 pKa = 4.34AYY958 pKa = 10.57DD959 pKa = 3.57TGLTTDD965 pKa = 2.91RR966 pKa = 11.84WLVAGLTADD975 pKa = 3.78AGTVLINSTTGLNLDD990 pKa = 4.25LRR992 pKa = 11.84SNSDD996 pKa = 3.21GNEE999 pKa = 4.04VTSCTLPATPVILAANEE1016 pKa = 4.34GSVTPAVGATSAALLDD1032 pKa = 4.1ADD1034 pKa = 3.85YY1035 pKa = 8.08TAPGFATGDD1044 pKa = 3.79TVTVTVDD1051 pKa = 3.47AEE1053 pKa = 4.31VLTVDD1058 pKa = 4.44FTVEE1062 pKa = 4.18LPVDD1066 pKa = 4.06SIFNDD1071 pKa = 3.35RR1072 pKa = 11.84FEE1074 pKa = 4.09NN1075 pKa = 3.72
Molecular weight: 112.64 kDa
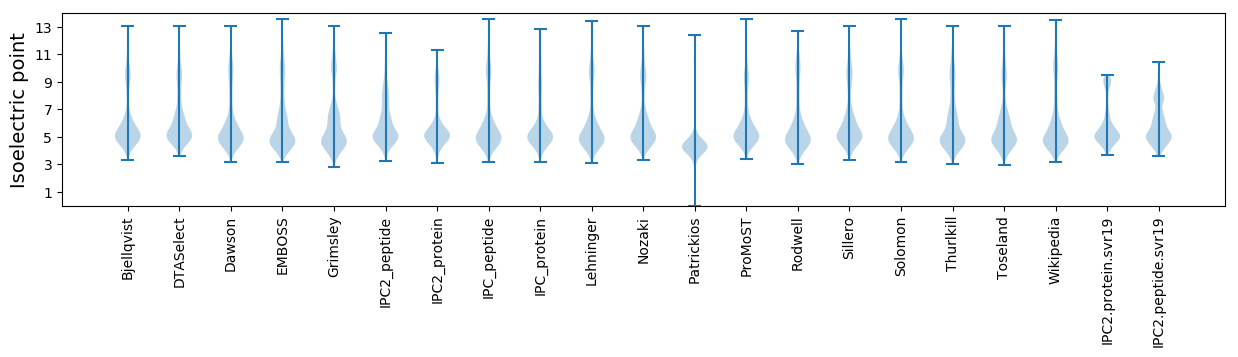
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K0XRS8|A0A0K0XRS8_9GAMM Glycine--tRNA ligase alpha subunit OS=Wenzhouxiangella marina OX=1579979 GN=glyQ PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84IKK11 pKa = 10.39RR12 pKa = 11.84VRR14 pKa = 11.84THH16 pKa = 5.87GFRR19 pKa = 11.84ARR21 pKa = 11.84MATASGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.81RR41 pKa = 11.84LAATNNRR48 pKa = 3.77
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84IKK11 pKa = 10.39RR12 pKa = 11.84VRR14 pKa = 11.84THH16 pKa = 5.87GFRR19 pKa = 11.84ARR21 pKa = 11.84MATASGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.81RR41 pKa = 11.84LAATNNRR48 pKa = 3.77
Molecular weight: 5.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1100236 |
29 |
5034 |
364.0 |
39.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.293 ± 0.061 | 0.888 ± 0.016 |
6.264 ± 0.039 | 6.606 ± 0.047 |
3.598 ± 0.028 | 8.554 ± 0.051 |
2.072 ± 0.026 | 4.746 ± 0.028 |
1.887 ± 0.032 | 11.339 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.107 ± 0.023 | 2.638 ± 0.041 |
5.217 ± 0.032 | 3.765 ± 0.025 |
7.827 ± 0.067 | 5.977 ± 0.047 |
4.588 ± 0.053 | 6.926 ± 0.043 |
1.573 ± 0.021 | 2.135 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
