
Eragrostis minor streak virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.92
Get precalculated fractions of proteins
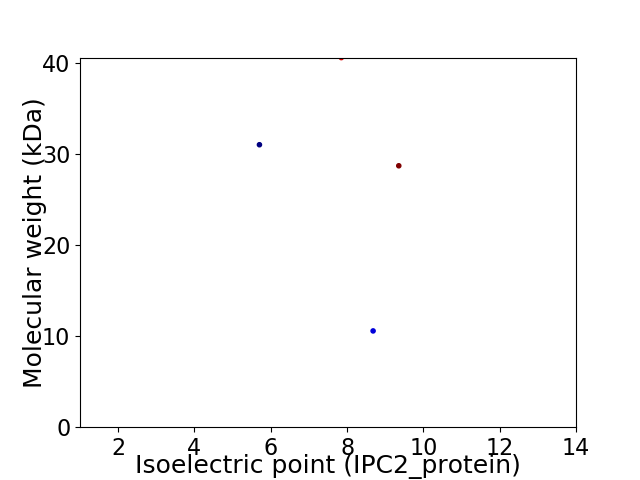
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
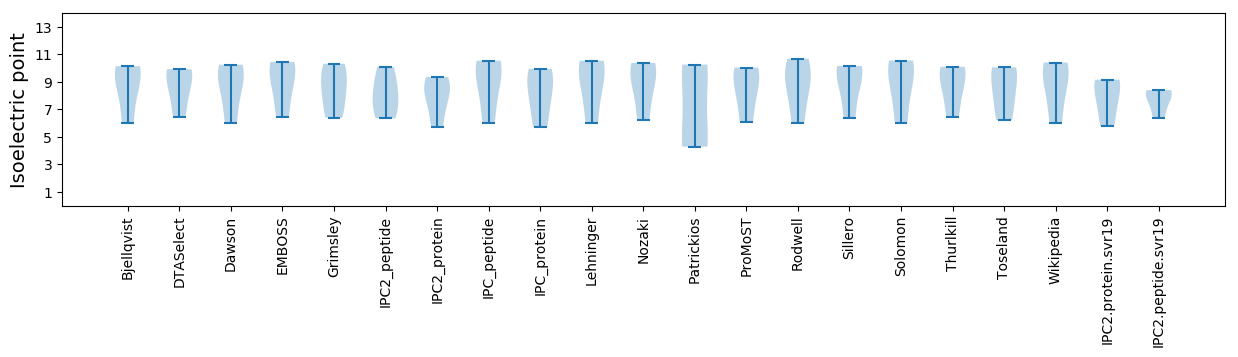
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F6M064|F6M064_9GEMI Replication-associated protein OS=Eragrostis minor streak virus OX=1030595 PE=3 SV=1
MM1 pKa = 7.3SQDD4 pKa = 3.14QDD6 pKa = 3.97TLGSSSDD13 pKa = 3.09GSRR16 pKa = 11.84FRR18 pKa = 11.84ISSKK22 pKa = 9.77QLFLTYY28 pKa = 9.42PRR30 pKa = 11.84CDD32 pKa = 4.15LSPKK36 pKa = 10.61DD37 pKa = 4.34LGLEE41 pKa = 3.98LLQLLIQNKK50 pKa = 8.73PKK52 pKa = 10.32YY53 pKa = 9.66IHH55 pKa = 5.73VTQEE59 pKa = 3.49LHH61 pKa = 7.14KK62 pKa = 11.11DD63 pKa = 4.03GFPHH67 pKa = 7.08LHH69 pKa = 6.84ALVQLEE75 pKa = 4.42KK76 pKa = 11.09KK77 pKa = 10.54LFTRR81 pKa = 11.84RR82 pKa = 11.84QTFFDD87 pKa = 3.65QFLHH91 pKa = 5.63GTKK94 pKa = 10.2FHH96 pKa = 7.18PNIQPARR103 pKa = 11.84DD104 pKa = 3.35ASKK107 pKa = 10.5VLGYY111 pKa = 8.45ITKK114 pKa = 10.44QNGEE118 pKa = 4.07EE119 pKa = 4.33YY120 pKa = 10.34IFGKK124 pKa = 8.31PTLPKK129 pKa = 10.06KK130 pKa = 10.63KK131 pKa = 8.99KK132 pKa = 7.45TAQEE136 pKa = 3.99GRR138 pKa = 11.84DD139 pKa = 3.28QRR141 pKa = 11.84MRR143 pKa = 11.84AIIEE147 pKa = 4.22SSTSKK152 pKa = 10.62QEE154 pKa = 3.88YY155 pKa = 9.13LSMVRR160 pKa = 11.84KK161 pKa = 9.06EE162 pKa = 5.3FPFDD166 pKa = 2.71WATRR170 pKa = 11.84LMQFEE175 pKa = 4.65YY176 pKa = 10.38SASSLFPEE184 pKa = 4.9PPVEE188 pKa = 4.18YY189 pKa = 9.74TSPFPVDD196 pKa = 3.54QLLCPEE202 pKa = 5.95DD203 pKa = 3.01ITEE206 pKa = 4.98IINSEE211 pKa = 4.0WFQVTPYY218 pKa = 10.26SYY220 pKa = 10.15TLIHH224 pKa = 7.22PGTSISQAADD234 pKa = 3.8DD235 pKa = 4.41LQQMQDD241 pKa = 2.19ISMEE245 pKa = 4.08HH246 pKa = 5.87QEE248 pKa = 4.3AGHH251 pKa = 5.64VASTSVDD258 pKa = 3.43QPVLEE263 pKa = 4.25RR264 pKa = 11.84RR265 pKa = 11.84LGLEE269 pKa = 3.8AA270 pKa = 5.82
MM1 pKa = 7.3SQDD4 pKa = 3.14QDD6 pKa = 3.97TLGSSSDD13 pKa = 3.09GSRR16 pKa = 11.84FRR18 pKa = 11.84ISSKK22 pKa = 9.77QLFLTYY28 pKa = 9.42PRR30 pKa = 11.84CDD32 pKa = 4.15LSPKK36 pKa = 10.61DD37 pKa = 4.34LGLEE41 pKa = 3.98LLQLLIQNKK50 pKa = 8.73PKK52 pKa = 10.32YY53 pKa = 9.66IHH55 pKa = 5.73VTQEE59 pKa = 3.49LHH61 pKa = 7.14KK62 pKa = 11.11DD63 pKa = 4.03GFPHH67 pKa = 7.08LHH69 pKa = 6.84ALVQLEE75 pKa = 4.42KK76 pKa = 11.09KK77 pKa = 10.54LFTRR81 pKa = 11.84RR82 pKa = 11.84QTFFDD87 pKa = 3.65QFLHH91 pKa = 5.63GTKK94 pKa = 10.2FHH96 pKa = 7.18PNIQPARR103 pKa = 11.84DD104 pKa = 3.35ASKK107 pKa = 10.5VLGYY111 pKa = 8.45ITKK114 pKa = 10.44QNGEE118 pKa = 4.07EE119 pKa = 4.33YY120 pKa = 10.34IFGKK124 pKa = 8.31PTLPKK129 pKa = 10.06KK130 pKa = 10.63KK131 pKa = 8.99KK132 pKa = 7.45TAQEE136 pKa = 3.99GRR138 pKa = 11.84DD139 pKa = 3.28QRR141 pKa = 11.84MRR143 pKa = 11.84AIIEE147 pKa = 4.22SSTSKK152 pKa = 10.62QEE154 pKa = 3.88YY155 pKa = 9.13LSMVRR160 pKa = 11.84KK161 pKa = 9.06EE162 pKa = 5.3FPFDD166 pKa = 2.71WATRR170 pKa = 11.84LMQFEE175 pKa = 4.65YY176 pKa = 10.38SASSLFPEE184 pKa = 4.9PPVEE188 pKa = 4.18YY189 pKa = 9.74TSPFPVDD196 pKa = 3.54QLLCPEE202 pKa = 5.95DD203 pKa = 3.01ITEE206 pKa = 4.98IINSEE211 pKa = 4.0WFQVTPYY218 pKa = 10.26SYY220 pKa = 10.15TLIHH224 pKa = 7.22PGTSISQAADD234 pKa = 3.8DD235 pKa = 4.41LQQMQDD241 pKa = 2.19ISMEE245 pKa = 4.08HH246 pKa = 5.87QEE248 pKa = 4.3AGHH251 pKa = 5.64VASTSVDD258 pKa = 3.43QPVLEE263 pKa = 4.25RR264 pKa = 11.84RR265 pKa = 11.84LGLEE269 pKa = 3.8AA270 pKa = 5.82
Molecular weight: 31.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F6M063|F6M063_9GEMI Replication-associated protein OS=Eragrostis minor streak virus OX=1030595 PE=3 SV=1
MM1 pKa = 7.97VYY3 pKa = 9.89EE4 pKa = 5.22RR5 pKa = 11.84KK6 pKa = 10.17RR7 pKa = 11.84KK8 pKa = 9.63DD9 pKa = 3.11PRR11 pKa = 11.84RR12 pKa = 11.84SDD14 pKa = 3.35EE15 pKa = 4.2SAGSARR21 pKa = 11.84KK22 pKa = 9.44RR23 pKa = 11.84PRR25 pKa = 11.84APAGRR30 pKa = 11.84SMVSAVRR37 pKa = 11.84RR38 pKa = 11.84PALQIRR44 pKa = 11.84EE45 pKa = 4.45YY46 pKa = 9.92PWQTLTQLPIKK57 pKa = 9.36ITDD60 pKa = 3.63GVIFMVNTIDD70 pKa = 4.92PGTGDD75 pKa = 3.78DD76 pKa = 4.61QRR78 pKa = 11.84SKK80 pKa = 10.43HH81 pKa = 4.41QTMLYY86 pKa = 10.32KK87 pKa = 10.1MSFNCVLWPDD97 pKa = 3.54EE98 pKa = 4.2TTALIVAPFRR108 pKa = 11.84VNFWLVYY115 pKa = 10.26DD116 pKa = 4.84AAPQGKK122 pKa = 9.2LPALSDD128 pKa = 3.55IFSVPYY134 pKa = 9.33TKK136 pKa = 9.78WGNTWNVSRR145 pKa = 11.84TNVHH149 pKa = 5.71RR150 pKa = 11.84FVVKK154 pKa = 10.39RR155 pKa = 11.84KK156 pKa = 6.82WHH158 pKa = 5.36VDD160 pKa = 3.4YY161 pKa = 11.08NSSGTAVGKK170 pKa = 9.85KK171 pKa = 8.14QSPGSEE177 pKa = 4.04DD178 pKa = 3.92FPVKK182 pKa = 10.68NVVEE186 pKa = 4.28CNKK189 pKa = 9.57FFEE192 pKa = 4.32KK193 pKa = 10.73LRR195 pKa = 11.84VKK197 pKa = 9.34TEE199 pKa = 3.76WLNTTDD205 pKa = 3.5GTIGSVKK212 pKa = 10.21KK213 pKa = 10.32GALYY217 pKa = 10.53LVANTRR223 pKa = 11.84QMPAGDD229 pKa = 4.51AVTTTCTTYY238 pKa = 9.54MQGSTRR244 pKa = 11.84LYY246 pKa = 9.94FKK248 pKa = 11.0SLGYY252 pKa = 9.73QQ253 pKa = 3.03
MM1 pKa = 7.97VYY3 pKa = 9.89EE4 pKa = 5.22RR5 pKa = 11.84KK6 pKa = 10.17RR7 pKa = 11.84KK8 pKa = 9.63DD9 pKa = 3.11PRR11 pKa = 11.84RR12 pKa = 11.84SDD14 pKa = 3.35EE15 pKa = 4.2SAGSARR21 pKa = 11.84KK22 pKa = 9.44RR23 pKa = 11.84PRR25 pKa = 11.84APAGRR30 pKa = 11.84SMVSAVRR37 pKa = 11.84RR38 pKa = 11.84PALQIRR44 pKa = 11.84EE45 pKa = 4.45YY46 pKa = 9.92PWQTLTQLPIKK57 pKa = 9.36ITDD60 pKa = 3.63GVIFMVNTIDD70 pKa = 4.92PGTGDD75 pKa = 3.78DD76 pKa = 4.61QRR78 pKa = 11.84SKK80 pKa = 10.43HH81 pKa = 4.41QTMLYY86 pKa = 10.32KK87 pKa = 10.1MSFNCVLWPDD97 pKa = 3.54EE98 pKa = 4.2TTALIVAPFRR108 pKa = 11.84VNFWLVYY115 pKa = 10.26DD116 pKa = 4.84AAPQGKK122 pKa = 9.2LPALSDD128 pKa = 3.55IFSVPYY134 pKa = 9.33TKK136 pKa = 9.78WGNTWNVSRR145 pKa = 11.84TNVHH149 pKa = 5.71RR150 pKa = 11.84FVVKK154 pKa = 10.39RR155 pKa = 11.84KK156 pKa = 6.82WHH158 pKa = 5.36VDD160 pKa = 3.4YY161 pKa = 11.08NSSGTAVGKK170 pKa = 9.85KK171 pKa = 8.14QSPGSEE177 pKa = 4.04DD178 pKa = 3.92FPVKK182 pKa = 10.68NVVEE186 pKa = 4.28CNKK189 pKa = 9.57FFEE192 pKa = 4.32KK193 pKa = 10.73LRR195 pKa = 11.84VKK197 pKa = 9.34TEE199 pKa = 3.76WLNTTDD205 pKa = 3.5GTIGSVKK212 pKa = 10.21KK213 pKa = 10.32GALYY217 pKa = 10.53LVANTRR223 pKa = 11.84QMPAGDD229 pKa = 4.51AVTTTCTTYY238 pKa = 9.54MQGSTRR244 pKa = 11.84LYY246 pKa = 9.94FKK248 pKa = 11.0SLGYY252 pKa = 9.73QQ253 pKa = 3.03
Molecular weight: 28.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
974 |
99 |
352 |
243.5 |
27.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.057 ± 1.064 | 1.129 ± 0.156 |
5.339 ± 0.509 | 5.441 ± 0.725 |
4.928 ± 0.295 | 5.749 ± 0.408 |
2.567 ± 0.55 | 4.723 ± 0.474 |
6.674 ± 1.006 | 8.932 ± 1.096 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.053 ± 0.256 | 2.567 ± 0.585 |
6.982 ± 0.575 | 6.057 ± 0.877 |
6.057 ± 0.855 | 8.008 ± 0.97 |
6.263 ± 0.955 | 5.031 ± 1.177 |
1.54 ± 0.365 | 3.901 ± 0.73 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
