
Escherichia coli (strain K12)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Escherichia; Escherichia coli
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
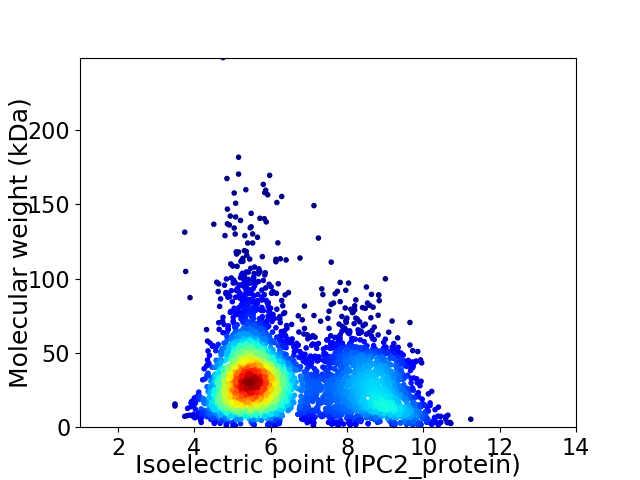
Virtual 2D-PAGE plot for 4450 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P77263|ECPE_ECOLI Probable fimbrial chaperone EcpE OS=Escherichia coli (strain K12) OX=83333 GN=ecpE PE=3 SV=2
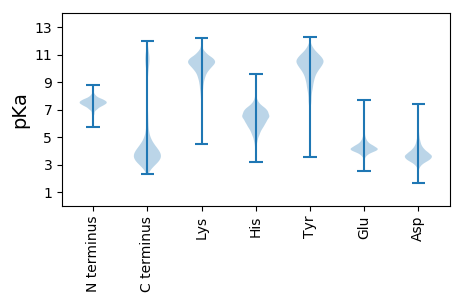
MM1 pKa = 7.37HH2 pKa = 6.88SWKK5 pKa = 10.29KK6 pKa = 10.49KK7 pKa = 9.88LVVSQLALACTLAITSQANAANYY30 pKa = 7.61DD31 pKa = 3.01TWTYY35 pKa = 10.78IDD37 pKa = 4.17NPVTALDD44 pKa = 3.82WDD46 pKa = 4.14HH47 pKa = 6.59MDD49 pKa = 4.13KK50 pKa = 11.31AGTVDD55 pKa = 3.75GNYY58 pKa = 9.86VNYY61 pKa = 10.78SGFVYY66 pKa = 10.79YY67 pKa = 11.15NNTNGDD73 pKa = 3.55FDD75 pKa = 3.99QSFNGDD81 pKa = 3.76TVNGTISTYY90 pKa = 10.77YY91 pKa = 10.45LNHH94 pKa = 7.64DD95 pKa = 4.38YY96 pKa = 11.55ADD98 pKa = 3.63STANQLDD105 pKa = 3.34ISNSVIHH112 pKa = 6.67GSITSMLPGGYY123 pKa = 9.26YY124 pKa = 10.45DD125 pKa = 5.9RR126 pKa = 11.84FDD128 pKa = 5.03ADD130 pKa = 3.75GNNLGGYY137 pKa = 9.86DD138 pKa = 4.6FYY140 pKa = 11.33TDD142 pKa = 3.91AVVDD146 pKa = 3.73THH148 pKa = 6.51WRR150 pKa = 11.84DD151 pKa = 3.02GDD153 pKa = 3.99VFTLNIANTTIDD165 pKa = 3.87DD166 pKa = 4.5DD167 pKa = 4.7YY168 pKa = 11.2EE169 pKa = 4.78ALYY172 pKa = 8.53FTDD175 pKa = 3.76SYY177 pKa = 11.73KK178 pKa = 11.25DD179 pKa = 3.37GDD181 pKa = 4.13VTKK184 pKa = 9.29HH185 pKa = 4.83TNEE188 pKa = 4.07TFDD191 pKa = 3.28TSEE194 pKa = 4.05GVAVNLDD201 pKa = 3.37VEE203 pKa = 4.96SNINISNNSRR213 pKa = 11.84VAGIALSQGNTYY225 pKa = 10.89NEE227 pKa = 4.58TYY229 pKa = 7.94TTEE232 pKa = 3.61SHH234 pKa = 5.81TWDD237 pKa = 3.34NNISVKK243 pKa = 10.76DD244 pKa = 3.58STVTSGSNYY253 pKa = 9.61ILDD256 pKa = 3.76SNTYY260 pKa = 9.99GKK262 pKa = 8.47TGHH265 pKa = 6.7FGNSDD270 pKa = 3.73EE271 pKa = 4.67PSDD274 pKa = 3.87YY275 pKa = 11.32AGPGDD280 pKa = 3.52VAMSFTASGSDD291 pKa = 3.35YY292 pKa = 11.64AMKK295 pKa = 11.03NNVFLSNSTLMGDD308 pKa = 3.89VAFTSTWNSNFDD320 pKa = 4.29PNGHH324 pKa = 7.08DD325 pKa = 3.76SNGDD329 pKa = 3.44GVKK332 pKa = 9.45DD333 pKa = 3.94TNGGWTDD340 pKa = 3.46DD341 pKa = 3.73SLNVDD346 pKa = 4.39EE347 pKa = 6.58LNLTLDD353 pKa = 4.31NGSKK357 pKa = 9.66WVGQAIYY364 pKa = 10.76NVAEE368 pKa = 4.05TSAMYY373 pKa = 10.4DD374 pKa = 3.21VATNSLTPDD383 pKa = 2.86ATYY386 pKa = 11.15EE387 pKa = 4.0NNDD390 pKa = 3.16WKK392 pKa = 10.85RR393 pKa = 11.84VVDD396 pKa = 4.29DD397 pKa = 5.09KK398 pKa = 11.71VFQSGVFNVALNNGSEE414 pKa = 3.96WDD416 pKa = 3.5TTGRR420 pKa = 11.84SIVDD424 pKa = 3.31TLTVNNGSQVNVSEE438 pKa = 4.74SKK440 pKa = 9.7LTSDD444 pKa = 5.45TIDD447 pKa = 3.5LTNGSSLNIGEE458 pKa = 4.88DD459 pKa = 4.23GYY461 pKa = 11.93VDD463 pKa = 3.86TDD465 pKa = 3.34HH466 pKa = 6.86LTINSYY472 pKa = 8.68STVALTEE479 pKa = 4.2STGWGADD486 pKa = 3.35YY487 pKa = 11.23NLYY490 pKa = 11.06ANTITVTNGGVLDD503 pKa = 4.08VNVDD507 pKa = 3.57QFDD510 pKa = 4.01TEE512 pKa = 4.09AFRR515 pKa = 11.84TDD517 pKa = 3.3KK518 pKa = 11.63LEE520 pKa = 4.15LTSGNIADD528 pKa = 4.0HH529 pKa = 6.51NGNVVSGVFDD539 pKa = 3.21IHH541 pKa = 8.66SSDD544 pKa = 3.72YY545 pKa = 11.19VLNADD550 pKa = 4.92LVNDD554 pKa = 4.26RR555 pKa = 11.84TWDD558 pKa = 3.61TSKK561 pKa = 11.36SNYY564 pKa = 9.88GYY566 pKa = 10.9GIVAMNSDD574 pKa = 3.53GHH576 pKa = 5.81LTINGNGDD584 pKa = 3.17VDD586 pKa = 4.69NGTEE590 pKa = 4.24LDD592 pKa = 3.71NSSVDD597 pKa = 3.52NVVAATGNYY606 pKa = 8.96KK607 pKa = 10.02VRR609 pKa = 11.84IDD611 pKa = 3.53NATGAGAIADD621 pKa = 4.08YY622 pKa = 10.27KK623 pKa = 10.72DD624 pKa = 3.46KK625 pKa = 11.16EE626 pKa = 4.21IIYY629 pKa = 10.79VNDD632 pKa = 3.27VNSNATFSAANKK644 pKa = 9.86ADD646 pKa = 3.72LGAYY650 pKa = 7.69TYY652 pKa = 10.29QAEE655 pKa = 4.36QRR657 pKa = 11.84GNTVVLQQMEE667 pKa = 4.34LTDD670 pKa = 4.16YY671 pKa = 11.84ANMALSIPSANTNIWNLEE689 pKa = 3.6QDD691 pKa = 3.86TVGTRR696 pKa = 11.84LTNSRR701 pKa = 11.84HH702 pKa = 4.84GLADD706 pKa = 3.5NGGAWVSYY714 pKa = 10.43FGGNFNGDD722 pKa = 3.55NGTINYY728 pKa = 8.82DD729 pKa = 3.13QDD731 pKa = 3.61VNGIMVGVDD740 pKa = 2.97TKK742 pKa = 11.02IDD744 pKa = 3.49GNNAKK749 pKa = 9.7WIVGAAAGFAKK760 pKa = 10.62GDD762 pKa = 3.55MNDD765 pKa = 3.31RR766 pKa = 11.84SGQVDD771 pKa = 3.54QDD773 pKa = 3.63SQTAYY778 pKa = 9.18IYY780 pKa = 11.0SSAHH784 pKa = 5.1FANNVFVDD792 pKa = 4.71GSLSYY797 pKa = 11.41SHH799 pKa = 7.31FNNDD803 pKa = 3.89LSATMSNGTYY813 pKa = 10.5VDD815 pKa = 3.75GSTNSDD821 pKa = 2.36AWGFGLKK828 pKa = 10.12AGYY831 pKa = 9.72DD832 pKa = 3.82FKK834 pKa = 11.38LGDD837 pKa = 3.77AGYY840 pKa = 7.83VTPYY844 pKa = 10.65GSVSGLFQSGDD855 pKa = 3.79DD856 pKa = 3.89YY857 pKa = 11.83QLSNDD862 pKa = 3.96MKK864 pKa = 11.23VDD866 pKa = 3.62GQSYY870 pKa = 10.64DD871 pKa = 3.02SMRR874 pKa = 11.84YY875 pKa = 8.94EE876 pKa = 4.34LGVDD880 pKa = 3.21AGYY883 pKa = 8.53TFTYY887 pKa = 10.6SEE889 pKa = 4.47DD890 pKa = 3.66QALTPYY896 pKa = 10.37FKK898 pKa = 10.64LAYY901 pKa = 10.52VYY903 pKa = 11.0DD904 pKa = 4.62DD905 pKa = 4.36SNNDD909 pKa = 2.94NDD911 pKa = 4.18VNGDD915 pKa = 3.89SIDD918 pKa = 3.64NGTEE922 pKa = 3.72GSAVRR927 pKa = 11.84VGLGTQFSFTKK938 pKa = 10.49NFSAYY943 pKa = 9.3TDD945 pKa = 3.45ANYY948 pKa = 10.81LGGGDD953 pKa = 4.6VDD955 pKa = 4.32QDD957 pKa = 2.67WSANVGVKK965 pKa = 7.43YY966 pKa = 8.84TWW968 pKa = 3.09
MM1 pKa = 7.37HH2 pKa = 6.88SWKK5 pKa = 10.29KK6 pKa = 10.49KK7 pKa = 9.88LVVSQLALACTLAITSQANAANYY30 pKa = 7.61DD31 pKa = 3.01TWTYY35 pKa = 10.78IDD37 pKa = 4.17NPVTALDD44 pKa = 3.82WDD46 pKa = 4.14HH47 pKa = 6.59MDD49 pKa = 4.13KK50 pKa = 11.31AGTVDD55 pKa = 3.75GNYY58 pKa = 9.86VNYY61 pKa = 10.78SGFVYY66 pKa = 10.79YY67 pKa = 11.15NNTNGDD73 pKa = 3.55FDD75 pKa = 3.99QSFNGDD81 pKa = 3.76TVNGTISTYY90 pKa = 10.77YY91 pKa = 10.45LNHH94 pKa = 7.64DD95 pKa = 4.38YY96 pKa = 11.55ADD98 pKa = 3.63STANQLDD105 pKa = 3.34ISNSVIHH112 pKa = 6.67GSITSMLPGGYY123 pKa = 9.26YY124 pKa = 10.45DD125 pKa = 5.9RR126 pKa = 11.84FDD128 pKa = 5.03ADD130 pKa = 3.75GNNLGGYY137 pKa = 9.86DD138 pKa = 4.6FYY140 pKa = 11.33TDD142 pKa = 3.91AVVDD146 pKa = 3.73THH148 pKa = 6.51WRR150 pKa = 11.84DD151 pKa = 3.02GDD153 pKa = 3.99VFTLNIANTTIDD165 pKa = 3.87DD166 pKa = 4.5DD167 pKa = 4.7YY168 pKa = 11.2EE169 pKa = 4.78ALYY172 pKa = 8.53FTDD175 pKa = 3.76SYY177 pKa = 11.73KK178 pKa = 11.25DD179 pKa = 3.37GDD181 pKa = 4.13VTKK184 pKa = 9.29HH185 pKa = 4.83TNEE188 pKa = 4.07TFDD191 pKa = 3.28TSEE194 pKa = 4.05GVAVNLDD201 pKa = 3.37VEE203 pKa = 4.96SNINISNNSRR213 pKa = 11.84VAGIALSQGNTYY225 pKa = 10.89NEE227 pKa = 4.58TYY229 pKa = 7.94TTEE232 pKa = 3.61SHH234 pKa = 5.81TWDD237 pKa = 3.34NNISVKK243 pKa = 10.76DD244 pKa = 3.58STVTSGSNYY253 pKa = 9.61ILDD256 pKa = 3.76SNTYY260 pKa = 9.99GKK262 pKa = 8.47TGHH265 pKa = 6.7FGNSDD270 pKa = 3.73EE271 pKa = 4.67PSDD274 pKa = 3.87YY275 pKa = 11.32AGPGDD280 pKa = 3.52VAMSFTASGSDD291 pKa = 3.35YY292 pKa = 11.64AMKK295 pKa = 11.03NNVFLSNSTLMGDD308 pKa = 3.89VAFTSTWNSNFDD320 pKa = 4.29PNGHH324 pKa = 7.08DD325 pKa = 3.76SNGDD329 pKa = 3.44GVKK332 pKa = 9.45DD333 pKa = 3.94TNGGWTDD340 pKa = 3.46DD341 pKa = 3.73SLNVDD346 pKa = 4.39EE347 pKa = 6.58LNLTLDD353 pKa = 4.31NGSKK357 pKa = 9.66WVGQAIYY364 pKa = 10.76NVAEE368 pKa = 4.05TSAMYY373 pKa = 10.4DD374 pKa = 3.21VATNSLTPDD383 pKa = 2.86ATYY386 pKa = 11.15EE387 pKa = 4.0NNDD390 pKa = 3.16WKK392 pKa = 10.85RR393 pKa = 11.84VVDD396 pKa = 4.29DD397 pKa = 5.09KK398 pKa = 11.71VFQSGVFNVALNNGSEE414 pKa = 3.96WDD416 pKa = 3.5TTGRR420 pKa = 11.84SIVDD424 pKa = 3.31TLTVNNGSQVNVSEE438 pKa = 4.74SKK440 pKa = 9.7LTSDD444 pKa = 5.45TIDD447 pKa = 3.5LTNGSSLNIGEE458 pKa = 4.88DD459 pKa = 4.23GYY461 pKa = 11.93VDD463 pKa = 3.86TDD465 pKa = 3.34HH466 pKa = 6.86LTINSYY472 pKa = 8.68STVALTEE479 pKa = 4.2STGWGADD486 pKa = 3.35YY487 pKa = 11.23NLYY490 pKa = 11.06ANTITVTNGGVLDD503 pKa = 4.08VNVDD507 pKa = 3.57QFDD510 pKa = 4.01TEE512 pKa = 4.09AFRR515 pKa = 11.84TDD517 pKa = 3.3KK518 pKa = 11.63LEE520 pKa = 4.15LTSGNIADD528 pKa = 4.0HH529 pKa = 6.51NGNVVSGVFDD539 pKa = 3.21IHH541 pKa = 8.66SSDD544 pKa = 3.72YY545 pKa = 11.19VLNADD550 pKa = 4.92LVNDD554 pKa = 4.26RR555 pKa = 11.84TWDD558 pKa = 3.61TSKK561 pKa = 11.36SNYY564 pKa = 9.88GYY566 pKa = 10.9GIVAMNSDD574 pKa = 3.53GHH576 pKa = 5.81LTINGNGDD584 pKa = 3.17VDD586 pKa = 4.69NGTEE590 pKa = 4.24LDD592 pKa = 3.71NSSVDD597 pKa = 3.52NVVAATGNYY606 pKa = 8.96KK607 pKa = 10.02VRR609 pKa = 11.84IDD611 pKa = 3.53NATGAGAIADD621 pKa = 4.08YY622 pKa = 10.27KK623 pKa = 10.72DD624 pKa = 3.46KK625 pKa = 11.16EE626 pKa = 4.21IIYY629 pKa = 10.79VNDD632 pKa = 3.27VNSNATFSAANKK644 pKa = 9.86ADD646 pKa = 3.72LGAYY650 pKa = 7.69TYY652 pKa = 10.29QAEE655 pKa = 4.36QRR657 pKa = 11.84GNTVVLQQMEE667 pKa = 4.34LTDD670 pKa = 4.16YY671 pKa = 11.84ANMALSIPSANTNIWNLEE689 pKa = 3.6QDD691 pKa = 3.86TVGTRR696 pKa = 11.84LTNSRR701 pKa = 11.84HH702 pKa = 4.84GLADD706 pKa = 3.5NGGAWVSYY714 pKa = 10.43FGGNFNGDD722 pKa = 3.55NGTINYY728 pKa = 8.82DD729 pKa = 3.13QDD731 pKa = 3.61VNGIMVGVDD740 pKa = 2.97TKK742 pKa = 11.02IDD744 pKa = 3.49GNNAKK749 pKa = 9.7WIVGAAAGFAKK760 pKa = 10.62GDD762 pKa = 3.55MNDD765 pKa = 3.31RR766 pKa = 11.84SGQVDD771 pKa = 3.54QDD773 pKa = 3.63SQTAYY778 pKa = 9.18IYY780 pKa = 11.0SSAHH784 pKa = 5.1FANNVFVDD792 pKa = 4.71GSLSYY797 pKa = 11.41SHH799 pKa = 7.31FNNDD803 pKa = 3.89LSATMSNGTYY813 pKa = 10.5VDD815 pKa = 3.75GSTNSDD821 pKa = 2.36AWGFGLKK828 pKa = 10.12AGYY831 pKa = 9.72DD832 pKa = 3.82FKK834 pKa = 11.38LGDD837 pKa = 3.77AGYY840 pKa = 7.83VTPYY844 pKa = 10.65GSVSGLFQSGDD855 pKa = 3.79DD856 pKa = 3.89YY857 pKa = 11.83QLSNDD862 pKa = 3.96MKK864 pKa = 11.23VDD866 pKa = 3.62GQSYY870 pKa = 10.64DD871 pKa = 3.02SMRR874 pKa = 11.84YY875 pKa = 8.94EE876 pKa = 4.34LGVDD880 pKa = 3.21AGYY883 pKa = 8.53TFTYY887 pKa = 10.6SEE889 pKa = 4.47DD890 pKa = 3.66QALTPYY896 pKa = 10.37FKK898 pKa = 10.64LAYY901 pKa = 10.52VYY903 pKa = 11.0DD904 pKa = 4.62DD905 pKa = 4.36SNNDD909 pKa = 2.94NDD911 pKa = 4.18VNGDD915 pKa = 3.89SIDD918 pKa = 3.64NGTEE922 pKa = 3.72GSAVRR927 pKa = 11.84VGLGTQFSFTKK938 pKa = 10.49NFSAYY943 pKa = 9.3TDD945 pKa = 3.45ANYY948 pKa = 10.81LGGGDD953 pKa = 4.6VDD955 pKa = 4.32QDD957 pKa = 2.67WSANVGVKK965 pKa = 7.43YY966 pKa = 8.84TWW968 pKa = 3.09
Molecular weight: 104.72 kDa
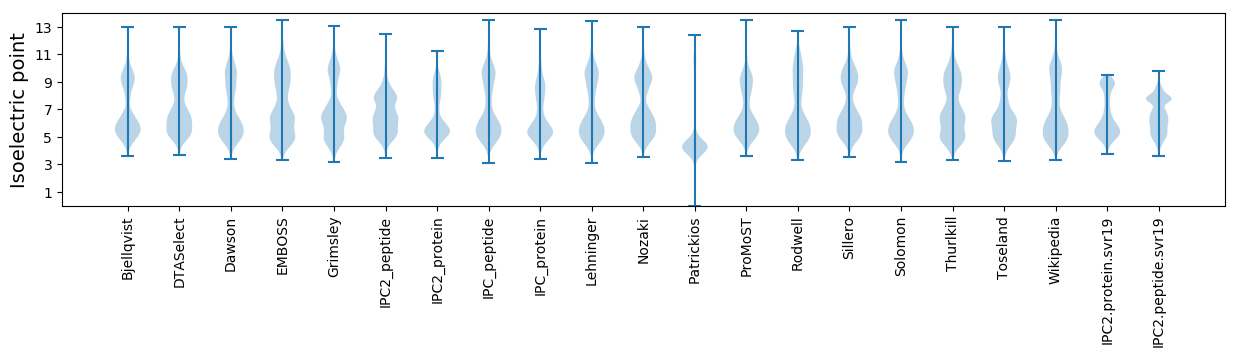
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P0A8K5|YAEP_ECOLI UPF0253 protein YaeP OS=Escherichia coli (strain K12) OX=83333 GN=yaeP PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LTVSKK46 pKa = 10.99
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.22GRR39 pKa = 11.84ARR41 pKa = 11.84LTVSKK46 pKa = 10.99
Molecular weight: 5.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1363428 |
8 |
2358 |
306.4 |
33.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.503 ± 0.048 | 1.162 ± 0.014 |
5.147 ± 0.026 | 5.77 ± 0.038 |
3.887 ± 0.026 | 7.36 ± 0.036 |
2.267 ± 0.022 | 6.011 ± 0.035 |
4.414 ± 0.034 | 10.671 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.828 ± 0.017 | 3.941 ± 0.029 |
4.428 ± 0.029 | 4.446 ± 0.03 |
5.532 ± 0.034 | 5.8 ± 0.03 |
5.389 ± 0.029 | 7.071 ± 0.028 |
1.526 ± 0.016 | 2.845 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
