
Gammapapillomavirus 16
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2D2AM05|A0A2D2AM05_9PAPI Minor capsid protein L2 OS=Gammapapillomavirus 16 OX=1513261 GN=L2 PE=3 SV=1
MM1 pKa = 7.35QGRR4 pKa = 11.84KK5 pKa = 5.03RR6 pKa = 11.84TKK8 pKa = 8.65RR9 pKa = 11.84ASVEE13 pKa = 3.94DD14 pKa = 4.57IYY16 pKa = 11.72AKK18 pKa = 10.8GCTQPGGYY26 pKa = 9.78CPPDD30 pKa = 3.29VKK32 pKa = 11.21NKK34 pKa = 10.1VEE36 pKa = 4.13GNTWADD42 pKa = 3.44WILKK46 pKa = 9.19VFGSVVYY53 pKa = 10.04FGGLGIGTGKK63 pKa = 8.78GTGGSTGYY71 pKa = 8.91TPLGGVGGSRR81 pKa = 11.84GPTTSIRR88 pKa = 11.84PTVPIEE94 pKa = 4.26TIGLPDD100 pKa = 3.67IVTVNPVEE108 pKa = 4.56PGASSIVPLAEE119 pKa = 4.28GLPEE123 pKa = 3.99PAVIDD128 pKa = 3.72TGSSIPGLAADD139 pKa = 4.11NTNITTVIDD148 pKa = 3.86PLSEE152 pKa = 3.98VTGLGEE158 pKa = 4.13HH159 pKa = 7.12PNLIGATSEE168 pKa = 4.04DD169 pKa = 3.97TVAVLDD175 pKa = 4.31VQTSPPPAKK184 pKa = 10.25KK185 pKa = 9.97ILLDD189 pKa = 3.56TSLTDD194 pKa = 3.01TTFRR198 pKa = 11.84VDD200 pKa = 2.89THH202 pKa = 6.75ASHH205 pKa = 7.36VDD207 pKa = 2.95ANLNIFVDD215 pKa = 3.95AQSFGSHH222 pKa = 6.29IGLSEE227 pKa = 5.26AIPLQEE233 pKa = 4.28INLISEE239 pKa = 4.42FEE241 pKa = 4.02IEE243 pKa = 4.27EE244 pKa = 5.1PIPSTSTPIAEE255 pKa = 4.04RR256 pKa = 11.84VVTRR260 pKa = 11.84AKK262 pKa = 10.21QLYY265 pKa = 8.21NRR267 pKa = 11.84YY268 pKa = 8.13VQQVPTRR275 pKa = 11.84PAEE278 pKa = 3.8FATYY282 pKa = 10.17SSRR285 pKa = 11.84FEE287 pKa = 4.22FEE289 pKa = 4.26NPAFMKK295 pKa = 10.57DD296 pKa = 3.18VTLEE300 pKa = 3.98FEE302 pKa = 4.17NDD304 pKa = 3.18LAEE307 pKa = 4.54IEE309 pKa = 4.83QITAPAISDD318 pKa = 3.35VRR320 pKa = 11.84ILNRR324 pKa = 11.84PTFSEE329 pKa = 4.27TTDD332 pKa = 3.06RR333 pKa = 11.84TVRR336 pKa = 11.84ISRR339 pKa = 11.84LGQRR343 pKa = 11.84AGMKK347 pKa = 8.73TRR349 pKa = 11.84SGLDD353 pKa = 2.89VGQRR357 pKa = 11.84VHH359 pKa = 7.29FYY361 pKa = 10.68YY362 pKa = 10.56DD363 pKa = 3.16VSKK366 pKa = 10.71ISSTIEE372 pKa = 3.5MNTFGEE378 pKa = 4.53YY379 pKa = 9.55SHH381 pKa = 7.06EE382 pKa = 4.1STIVDD387 pKa = 3.77EE388 pKa = 4.8LLSSSFINPFEE399 pKa = 4.14MPIEE403 pKa = 5.08DD404 pKa = 3.64IFSDD408 pKa = 3.95TEE410 pKa = 4.57LIDD413 pKa = 3.97PLEE416 pKa = 4.03EE417 pKa = 4.26DD418 pKa = 4.15FRR420 pKa = 11.84NSHH423 pKa = 7.21LILPFSDD430 pKa = 3.98DD431 pKa = 3.13QDD433 pKa = 4.14VYY435 pKa = 11.64ALPTLPPGIGLKK447 pKa = 10.5LYY449 pKa = 10.32TDD451 pKa = 3.98LTEE454 pKa = 4.71KK455 pKa = 10.85DD456 pKa = 4.06LFIHH460 pKa = 6.98HH461 pKa = 6.98PTQNTQIIVPDD472 pKa = 3.6SPYY475 pKa = 10.53IPLQPPIGAVIDD487 pKa = 3.6NDD489 pKa = 4.29DD490 pKa = 4.21YY491 pKa = 11.81YY492 pKa = 11.67LHH494 pKa = 7.46PSLLTRR500 pKa = 11.84KK501 pKa = 9.71RR502 pKa = 11.84KK503 pKa = 9.33RR504 pKa = 11.84AFFF507 pKa = 4.42
MM1 pKa = 7.35QGRR4 pKa = 11.84KK5 pKa = 5.03RR6 pKa = 11.84TKK8 pKa = 8.65RR9 pKa = 11.84ASVEE13 pKa = 3.94DD14 pKa = 4.57IYY16 pKa = 11.72AKK18 pKa = 10.8GCTQPGGYY26 pKa = 9.78CPPDD30 pKa = 3.29VKK32 pKa = 11.21NKK34 pKa = 10.1VEE36 pKa = 4.13GNTWADD42 pKa = 3.44WILKK46 pKa = 9.19VFGSVVYY53 pKa = 10.04FGGLGIGTGKK63 pKa = 8.78GTGGSTGYY71 pKa = 8.91TPLGGVGGSRR81 pKa = 11.84GPTTSIRR88 pKa = 11.84PTVPIEE94 pKa = 4.26TIGLPDD100 pKa = 3.67IVTVNPVEE108 pKa = 4.56PGASSIVPLAEE119 pKa = 4.28GLPEE123 pKa = 3.99PAVIDD128 pKa = 3.72TGSSIPGLAADD139 pKa = 4.11NTNITTVIDD148 pKa = 3.86PLSEE152 pKa = 3.98VTGLGEE158 pKa = 4.13HH159 pKa = 7.12PNLIGATSEE168 pKa = 4.04DD169 pKa = 3.97TVAVLDD175 pKa = 4.31VQTSPPPAKK184 pKa = 10.25KK185 pKa = 9.97ILLDD189 pKa = 3.56TSLTDD194 pKa = 3.01TTFRR198 pKa = 11.84VDD200 pKa = 2.89THH202 pKa = 6.75ASHH205 pKa = 7.36VDD207 pKa = 2.95ANLNIFVDD215 pKa = 3.95AQSFGSHH222 pKa = 6.29IGLSEE227 pKa = 5.26AIPLQEE233 pKa = 4.28INLISEE239 pKa = 4.42FEE241 pKa = 4.02IEE243 pKa = 4.27EE244 pKa = 5.1PIPSTSTPIAEE255 pKa = 4.04RR256 pKa = 11.84VVTRR260 pKa = 11.84AKK262 pKa = 10.21QLYY265 pKa = 8.21NRR267 pKa = 11.84YY268 pKa = 8.13VQQVPTRR275 pKa = 11.84PAEE278 pKa = 3.8FATYY282 pKa = 10.17SSRR285 pKa = 11.84FEE287 pKa = 4.22FEE289 pKa = 4.26NPAFMKK295 pKa = 10.57DD296 pKa = 3.18VTLEE300 pKa = 3.98FEE302 pKa = 4.17NDD304 pKa = 3.18LAEE307 pKa = 4.54IEE309 pKa = 4.83QITAPAISDD318 pKa = 3.35VRR320 pKa = 11.84ILNRR324 pKa = 11.84PTFSEE329 pKa = 4.27TTDD332 pKa = 3.06RR333 pKa = 11.84TVRR336 pKa = 11.84ISRR339 pKa = 11.84LGQRR343 pKa = 11.84AGMKK347 pKa = 8.73TRR349 pKa = 11.84SGLDD353 pKa = 2.89VGQRR357 pKa = 11.84VHH359 pKa = 7.29FYY361 pKa = 10.68YY362 pKa = 10.56DD363 pKa = 3.16VSKK366 pKa = 10.71ISSTIEE372 pKa = 3.5MNTFGEE378 pKa = 4.53YY379 pKa = 9.55SHH381 pKa = 7.06EE382 pKa = 4.1STIVDD387 pKa = 3.77EE388 pKa = 4.8LLSSSFINPFEE399 pKa = 4.14MPIEE403 pKa = 5.08DD404 pKa = 3.64IFSDD408 pKa = 3.95TEE410 pKa = 4.57LIDD413 pKa = 3.97PLEE416 pKa = 4.03EE417 pKa = 4.26DD418 pKa = 4.15FRR420 pKa = 11.84NSHH423 pKa = 7.21LILPFSDD430 pKa = 3.98DD431 pKa = 3.13QDD433 pKa = 4.14VYY435 pKa = 11.64ALPTLPPGIGLKK447 pKa = 10.5LYY449 pKa = 10.32TDD451 pKa = 3.98LTEE454 pKa = 4.71KK455 pKa = 10.85DD456 pKa = 4.06LFIHH460 pKa = 6.98HH461 pKa = 6.98PTQNTQIIVPDD472 pKa = 3.6SPYY475 pKa = 10.53IPLQPPIGAVIDD487 pKa = 3.6NDD489 pKa = 4.29DD490 pKa = 4.21YY491 pKa = 11.81YY492 pKa = 11.67LHH494 pKa = 7.46PSLLTRR500 pKa = 11.84KK501 pKa = 9.71RR502 pKa = 11.84KK503 pKa = 9.33RR504 pKa = 11.84AFFF507 pKa = 4.42
Molecular weight: 55.42 kDa
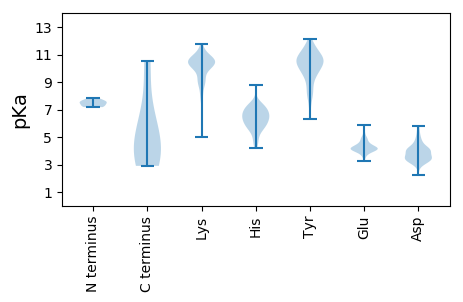
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2ALU7|A0A2D2ALU7_9PAPI Protein E7 OS=Gammapapillomavirus 16 OX=1513261 GN=E7 PE=3 SV=1
MM1 pKa = 7.6EE2 pKa = 5.86PLFPTKK8 pKa = 10.52LNEE11 pKa = 3.72YY12 pKa = 9.87CSYY15 pKa = 11.32FDD17 pKa = 4.62ISFFDD22 pKa = 4.64LSLKK26 pKa = 10.8CIFCKK31 pKa = 10.37HH32 pKa = 5.4YY33 pKa = 11.28LNLVDD38 pKa = 5.35LAYY41 pKa = 10.0FYY43 pKa = 11.23EE44 pKa = 5.27KK45 pKa = 10.7GLSLVWRR52 pKa = 11.84NNICYY57 pKa = 9.82ACCDD61 pKa = 3.41RR62 pKa = 11.84CLKK65 pKa = 10.72CSARR69 pKa = 11.84YY70 pKa = 7.05EE71 pKa = 4.36ANRR74 pKa = 11.84HH75 pKa = 4.58FQCTFNTATLHH86 pKa = 6.64AVLEE90 pKa = 4.54KK91 pKa = 10.1PLQDD95 pKa = 3.29IEE97 pKa = 4.14IRR99 pKa = 11.84CYY101 pKa = 10.45YY102 pKa = 9.24CLCILSLVEE111 pKa = 4.08KK112 pKa = 10.13CDD114 pKa = 4.3LIARR118 pKa = 11.84GRR120 pKa = 11.84PTCLVRR126 pKa = 11.84GYY128 pKa = 10.43FRR130 pKa = 11.84APCKK134 pKa = 10.39DD135 pKa = 3.55CIRR138 pKa = 11.84RR139 pKa = 11.84EE140 pKa = 3.95IYY142 pKa = 10.51
MM1 pKa = 7.6EE2 pKa = 5.86PLFPTKK8 pKa = 10.52LNEE11 pKa = 3.72YY12 pKa = 9.87CSYY15 pKa = 11.32FDD17 pKa = 4.62ISFFDD22 pKa = 4.64LSLKK26 pKa = 10.8CIFCKK31 pKa = 10.37HH32 pKa = 5.4YY33 pKa = 11.28LNLVDD38 pKa = 5.35LAYY41 pKa = 10.0FYY43 pKa = 11.23EE44 pKa = 5.27KK45 pKa = 10.7GLSLVWRR52 pKa = 11.84NNICYY57 pKa = 9.82ACCDD61 pKa = 3.41RR62 pKa = 11.84CLKK65 pKa = 10.72CSARR69 pKa = 11.84YY70 pKa = 7.05EE71 pKa = 4.36ANRR74 pKa = 11.84HH75 pKa = 4.58FQCTFNTATLHH86 pKa = 6.64AVLEE90 pKa = 4.54KK91 pKa = 10.1PLQDD95 pKa = 3.29IEE97 pKa = 4.14IRR99 pKa = 11.84CYY101 pKa = 10.45YY102 pKa = 9.24CLCILSLVEE111 pKa = 4.08KK112 pKa = 10.13CDD114 pKa = 4.3LIARR118 pKa = 11.84GRR120 pKa = 11.84PTCLVRR126 pKa = 11.84GYY128 pKa = 10.43FRR130 pKa = 11.84APCKK134 pKa = 10.39DD135 pKa = 3.55CIRR138 pKa = 11.84RR139 pKa = 11.84EE140 pKa = 3.95IYY142 pKa = 10.51
Molecular weight: 16.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2395 |
99 |
610 |
342.1 |
38.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.887 ± 0.251 | 2.38 ± 0.882 |
6.221 ± 0.586 | 6.681 ± 0.726 |
4.301 ± 0.548 | 5.261 ± 0.747 |
2.296 ± 0.239 | 5.428 ± 1.025 |
5.219 ± 0.679 | 9.812 ± 0.657 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.837 ± 0.361 | 5.762 ± 0.647 |
5.846 ± 1.012 | 3.967 ± 0.38 |
5.386 ± 0.823 | 7.307 ± 0.607 |
6.472 ± 0.784 | 5.01 ± 0.644 |
1.211 ± 0.296 | 3.716 ± 0.522 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
