
Chondromyces crocatus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Sorangiineae; Polyangiaceae; Chondromyces
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
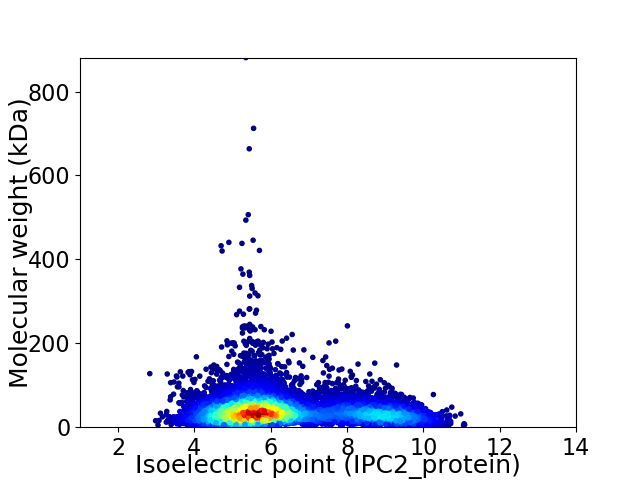
Virtual 2D-PAGE plot for 8327 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1EH87|A0A0K1EH87_CHOCO Uncharacterized protein OS=Chondromyces crocatus OX=52 GN=CMC5_041110 PE=4 SV=1
MM1 pKa = 7.42SRR3 pKa = 11.84FALKK7 pKa = 9.58MLGIAGVFGVFAACSATKK25 pKa = 10.67GGDD28 pKa = 3.23NSFGGSEE35 pKa = 4.28TTSSDD40 pKa = 2.92GTSQGGHH47 pKa = 5.99GAAGGDD53 pKa = 3.59SGEE56 pKa = 4.15GGIAIEE62 pKa = 4.41TGSGGGGSQPPCIDD76 pKa = 3.46TPGVDD81 pKa = 4.23DD82 pKa = 6.07DD83 pKa = 5.35GDD85 pKa = 3.86GFADD89 pKa = 4.79PEE91 pKa = 4.57DD92 pKa = 5.83CNDD95 pKa = 3.95CDD97 pKa = 4.74PNVNPAAMEE106 pKa = 4.04VMTAEE111 pKa = 4.4GADD114 pKa = 4.1PVDD117 pKa = 4.05EE118 pKa = 5.27DD119 pKa = 5.3CDD121 pKa = 4.57GEE123 pKa = 4.14IDD125 pKa = 4.89NIAGTCDD132 pKa = 3.21EE133 pKa = 4.6NLLVEE138 pKa = 5.5DD139 pKa = 5.51PDD141 pKa = 3.78PMSAVRR147 pKa = 11.84ALDD150 pKa = 3.0ICRR153 pKa = 11.84NAQGPRR159 pKa = 11.84DD160 pKa = 3.68WGLLGAAWVRR170 pKa = 11.84ANGTPLTAPTNQYY183 pKa = 10.67GIQPNFGPNVSPRR196 pKa = 11.84GGDD199 pKa = 2.9RR200 pKa = 11.84MLAISSGRR208 pKa = 11.84ARR210 pKa = 11.84LPGQTGACTTEE221 pKa = 3.87SCNGHH226 pKa = 6.26GLGTAPSGFPQSVTGCPVGSSINDD250 pKa = 3.42DD251 pKa = 3.37VGLQVHH257 pKa = 5.89MRR259 pKa = 11.84APSNATGLRR268 pKa = 11.84FNFRR272 pKa = 11.84FYY274 pKa = 11.01SFEE277 pKa = 4.0YY278 pKa = 10.44PEE280 pKa = 4.67FVCMSYY286 pKa = 11.26NDD288 pKa = 3.49QFIALMNPAPAGAANGNISFDD309 pKa = 3.58SMGNPVSVNVAFFDD323 pKa = 3.85VCEE326 pKa = 4.28GCPLGIADD334 pKa = 5.23LLGTGFDD341 pKa = 3.79TLDD344 pKa = 3.75DD345 pKa = 4.4AGATGWLATTAPVKK359 pKa = 10.56GGQEE363 pKa = 3.64VSLRR367 pKa = 11.84FAIWDD372 pKa = 3.93TGDD375 pKa = 3.22QQYY378 pKa = 11.6DD379 pKa = 3.49STALIDD385 pKa = 3.68NFEE388 pKa = 4.61WIATGGTVPVGTTPEE403 pKa = 4.06PQQ405 pKa = 2.9
MM1 pKa = 7.42SRR3 pKa = 11.84FALKK7 pKa = 9.58MLGIAGVFGVFAACSATKK25 pKa = 10.67GGDD28 pKa = 3.23NSFGGSEE35 pKa = 4.28TTSSDD40 pKa = 2.92GTSQGGHH47 pKa = 5.99GAAGGDD53 pKa = 3.59SGEE56 pKa = 4.15GGIAIEE62 pKa = 4.41TGSGGGGSQPPCIDD76 pKa = 3.46TPGVDD81 pKa = 4.23DD82 pKa = 6.07DD83 pKa = 5.35GDD85 pKa = 3.86GFADD89 pKa = 4.79PEE91 pKa = 4.57DD92 pKa = 5.83CNDD95 pKa = 3.95CDD97 pKa = 4.74PNVNPAAMEE106 pKa = 4.04VMTAEE111 pKa = 4.4GADD114 pKa = 4.1PVDD117 pKa = 4.05EE118 pKa = 5.27DD119 pKa = 5.3CDD121 pKa = 4.57GEE123 pKa = 4.14IDD125 pKa = 4.89NIAGTCDD132 pKa = 3.21EE133 pKa = 4.6NLLVEE138 pKa = 5.5DD139 pKa = 5.51PDD141 pKa = 3.78PMSAVRR147 pKa = 11.84ALDD150 pKa = 3.0ICRR153 pKa = 11.84NAQGPRR159 pKa = 11.84DD160 pKa = 3.68WGLLGAAWVRR170 pKa = 11.84ANGTPLTAPTNQYY183 pKa = 10.67GIQPNFGPNVSPRR196 pKa = 11.84GGDD199 pKa = 2.9RR200 pKa = 11.84MLAISSGRR208 pKa = 11.84ARR210 pKa = 11.84LPGQTGACTTEE221 pKa = 3.87SCNGHH226 pKa = 6.26GLGTAPSGFPQSVTGCPVGSSINDD250 pKa = 3.42DD251 pKa = 3.37VGLQVHH257 pKa = 5.89MRR259 pKa = 11.84APSNATGLRR268 pKa = 11.84FNFRR272 pKa = 11.84FYY274 pKa = 11.01SFEE277 pKa = 4.0YY278 pKa = 10.44PEE280 pKa = 4.67FVCMSYY286 pKa = 11.26NDD288 pKa = 3.49QFIALMNPAPAGAANGNISFDD309 pKa = 3.58SMGNPVSVNVAFFDD323 pKa = 3.85VCEE326 pKa = 4.28GCPLGIADD334 pKa = 5.23LLGTGFDD341 pKa = 3.79TLDD344 pKa = 3.75DD345 pKa = 4.4AGATGWLATTAPVKK359 pKa = 10.56GGQEE363 pKa = 3.64VSLRR367 pKa = 11.84FAIWDD372 pKa = 3.93TGDD375 pKa = 3.22QQYY378 pKa = 11.6DD379 pKa = 3.49STALIDD385 pKa = 3.68NFEE388 pKa = 4.61WIATGGTVPVGTTPEE403 pKa = 4.06PQQ405 pKa = 2.9
Molecular weight: 41.41 kDa
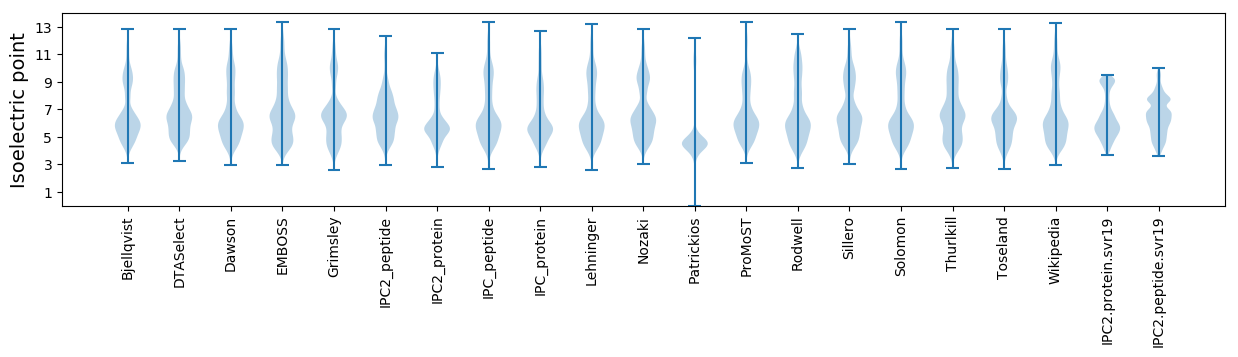
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1EEQ1|A0A0K1EEQ1_CHOCO Uncharacterized protein OS=Chondromyces crocatus OX=52 GN=CMC5_034760 PE=4 SV=1
MM1 pKa = 7.77LSGQFPVTRR10 pKa = 11.84SQFPVTRR17 pKa = 11.84SQFPVTRR24 pKa = 11.84SQFPVTRR31 pKa = 11.84SQFPVTRR38 pKa = 11.84SQFPVTRR45 pKa = 11.84SQFPVTRR52 pKa = 11.84SQFPVLSGQFPVTRR66 pKa = 11.84SQFPVVV72 pKa = 3.29
MM1 pKa = 7.77LSGQFPVTRR10 pKa = 11.84SQFPVTRR17 pKa = 11.84SQFPVTRR24 pKa = 11.84SQFPVTRR31 pKa = 11.84SQFPVTRR38 pKa = 11.84SQFPVTRR45 pKa = 11.84SQFPVTRR52 pKa = 11.84SQFPVLSGQFPVTRR66 pKa = 11.84SQFPVVV72 pKa = 3.29
Molecular weight: 8.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3359666 |
29 |
8158 |
403.5 |
43.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.72 ± 0.042 | 1.172 ± 0.018 |
5.547 ± 0.022 | 6.272 ± 0.028 |
3.042 ± 0.013 | 9.166 ± 0.042 |
2.201 ± 0.013 | 3.582 ± 0.018 |
2.389 ± 0.024 | 10.389 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.802 ± 0.012 | 1.9 ± 0.018 |
6.444 ± 0.026 | 3.083 ± 0.015 |
8.112 ± 0.032 | 5.899 ± 0.021 |
5.464 ± 0.023 | 7.645 ± 0.024 |
1.293 ± 0.009 | 1.879 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
