
Porcine epidemic diarrhea virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Alphacoronavirus; Pedacovirus
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
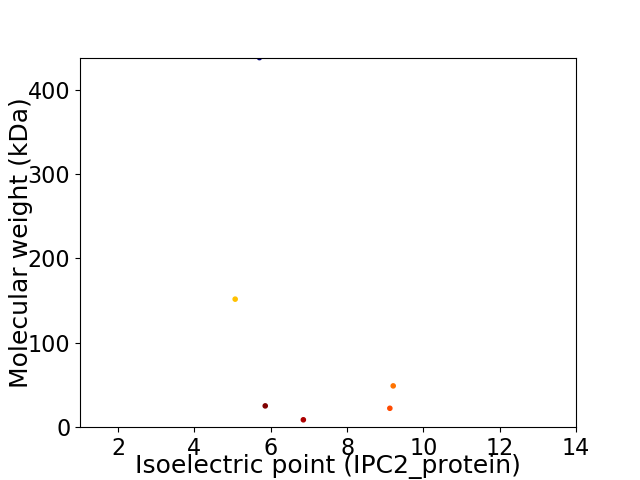
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
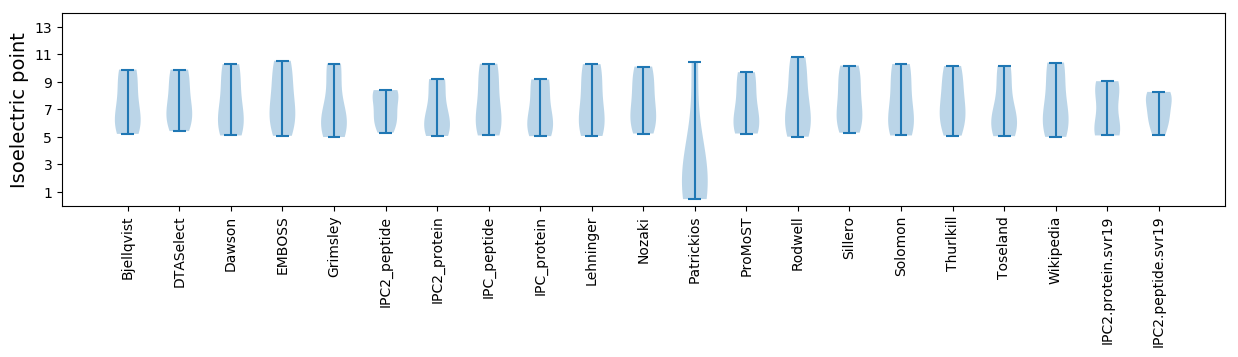
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A059VJE6|A0A059VJE6_9ALPC 3C-like proteinase OS=Porcine epidemic diarrhea virus OX=28295 GN=ORF1a PE=3 SV=1
MM1 pKa = 7.99KK2 pKa = 10.42SLTYY6 pKa = 9.83FWLFLPVLSTLSLPQDD22 pKa = 3.57VTRR25 pKa = 11.84CSANTNFRR33 pKa = 11.84RR34 pKa = 11.84FFSKK38 pKa = 10.72FNVQAPAVVVLGGYY52 pKa = 10.04LPIGEE57 pKa = 4.23NQGVNSTWYY66 pKa = 10.45CAGQHH71 pKa = 5.25PTASGVHH78 pKa = 6.7GIFVSHH84 pKa = 6.55IRR86 pKa = 11.84GGHH89 pKa = 4.59GFEE92 pKa = 4.58IGISQEE98 pKa = 3.85PFDD101 pKa = 4.04PSGYY105 pKa = 9.89QLYY108 pKa = 10.0LHH110 pKa = 7.06KK111 pKa = 10.35ATNGNTNATARR122 pKa = 11.84LRR124 pKa = 11.84ICQFPSIKK132 pKa = 9.87TLGPTANNDD141 pKa = 2.87VTTGRR146 pKa = 11.84NCLFNKK152 pKa = 9.98AIPAHH157 pKa = 5.62MSEE160 pKa = 4.46HH161 pKa = 5.85SVVGITWDD169 pKa = 3.48NDD171 pKa = 3.08RR172 pKa = 11.84VTVFSDD178 pKa = 3.69KK179 pKa = 10.46IYY181 pKa = 11.02YY182 pKa = 10.22FYY184 pKa = 10.98FKK186 pKa = 10.95NDD188 pKa = 3.08WSRR191 pKa = 11.84VATKK195 pKa = 10.01CYY197 pKa = 10.67NSGGCAMQYY206 pKa = 10.42VYY208 pKa = 11.04EE209 pKa = 4.08PTYY212 pKa = 11.64YY213 pKa = 9.87MLNVTSAGEE222 pKa = 4.14DD223 pKa = 3.74GISYY227 pKa = 9.02QPCTANCIGYY237 pKa = 9.17AANVFATEE245 pKa = 4.27PNGHH249 pKa = 6.21IPEE252 pKa = 4.55GFSFNNWFLLSNDD265 pKa = 3.05STLVHH270 pKa = 6.24GKK272 pKa = 9.44VVSNQPLLVNCLLAIPKK289 pKa = 9.74IYY291 pKa = 10.87GLGQFFSFNQTIDD304 pKa = 3.8GVCNGAAVQRR314 pKa = 11.84APEE317 pKa = 3.93ALRR320 pKa = 11.84FNINDD325 pKa = 3.42TSVILAEE332 pKa = 4.36GSIVLHH338 pKa = 6.0TALGTNFSFVCSNSSNPHH356 pKa = 6.09LATFAIPLGATQVPYY371 pKa = 11.25YY372 pKa = 10.63CFLKK376 pKa = 10.57VDD378 pKa = 4.51TYY380 pKa = 11.79NSTVYY385 pKa = 10.65KK386 pKa = 9.87FLAVLPPTVRR396 pKa = 11.84EE397 pKa = 3.97IVITKK402 pKa = 10.54YY403 pKa = 10.93GDD405 pKa = 3.33VYY407 pKa = 11.7VNGFGYY413 pKa = 10.69LHH415 pKa = 7.17LGLLDD420 pKa = 3.63AVTINFTGHH429 pKa = 5.73GTDD432 pKa = 4.59DD433 pKa = 4.08DD434 pKa = 5.06VSGFWTIASTNFVDD448 pKa = 5.9ALIEE452 pKa = 4.16VQGTAIQRR460 pKa = 11.84ILYY463 pKa = 10.06CDD465 pKa = 4.15DD466 pKa = 4.14PVSQLKK472 pKa = 10.32CSQVAFDD479 pKa = 5.4LDD481 pKa = 3.89DD482 pKa = 3.81GFYY485 pKa = 10.38PISSRR490 pKa = 11.84NLLSHH495 pKa = 6.55EE496 pKa = 4.12QPISFVTLPSFNDD509 pKa = 3.26HH510 pKa = 6.27SFVNITVSASFGGHH524 pKa = 5.66SGANLIASDD533 pKa = 3.51TTINGFSSFCVDD545 pKa = 2.78TRR547 pKa = 11.84QFTISLFYY555 pKa = 11.08NVTNSYY561 pKa = 11.02GYY563 pKa = 10.2VSKK566 pKa = 10.92SQDD569 pKa = 3.46SNCPFTLQSVNDD581 pKa = 3.71YY582 pKa = 11.36LSFSKK587 pKa = 10.79FCVSTSLLASACTIDD602 pKa = 4.13LFGYY606 pKa = 9.44PEE608 pKa = 4.41FGSGVKK614 pKa = 8.38FTSLYY619 pKa = 10.0FQFTKK624 pKa = 11.13GEE626 pKa = 4.85LITGTPKK633 pKa = 10.48PLEE636 pKa = 4.31GVTDD640 pKa = 3.65VSFMTLDD647 pKa = 3.0VCTKK651 pKa = 7.34YY652 pKa = 9.42TIYY655 pKa = 10.66GFKK658 pKa = 10.88GEE660 pKa = 4.78GIITLTNSSFLAGVYY675 pKa = 7.31YY676 pKa = 10.55TSDD679 pKa = 3.25SGQLLAFKK687 pKa = 10.21NVTSGAVYY695 pKa = 10.52SVTPCSFSEE704 pKa = 3.9QAAYY708 pKa = 10.85VDD710 pKa = 4.13DD711 pKa = 6.26DD712 pKa = 3.81IVGVISSLSSSTFNSTRR729 pKa = 11.84EE730 pKa = 3.91LPGFFYY736 pKa = 10.78HH737 pKa = 7.56SNDD740 pKa = 2.92GSNCTEE746 pKa = 3.82PVLVYY751 pKa = 11.07SNIGVCKK758 pKa = 9.81SGSIGYY764 pKa = 9.24VPSQSGQVKK773 pKa = 9.94IAPTVTGNISIPTNFSMSIRR793 pKa = 11.84TEE795 pKa = 3.69YY796 pKa = 10.03LQLYY800 pKa = 6.15NTPVSVDD807 pKa = 3.13CATYY811 pKa = 9.95VCNGNSRR818 pKa = 11.84CKK820 pKa = 10.28QLLTQYY826 pKa = 9.3TAACKK831 pKa = 9.6TIEE834 pKa = 4.39SALQLSARR842 pKa = 11.84LEE844 pKa = 4.32SVEE847 pKa = 4.22VNSMLTISEE856 pKa = 4.32EE857 pKa = 4.08ALQLATISSFNGDD870 pKa = 3.29GYY872 pKa = 11.6NFTNVLGVSVYY883 pKa = 10.82DD884 pKa = 3.71PASGRR889 pKa = 11.84VVQKK893 pKa = 10.67RR894 pKa = 11.84SFIEE898 pKa = 4.23DD899 pKa = 3.54LLFNKK904 pKa = 9.39VVTNGLGTVDD914 pKa = 4.28EE915 pKa = 4.8DD916 pKa = 4.8YY917 pKa = 11.12KK918 pKa = 10.79RR919 pKa = 11.84CSNGRR924 pKa = 11.84SVADD928 pKa = 3.65LVCAQYY934 pKa = 11.29YY935 pKa = 9.11SGVMVLPGVVDD946 pKa = 3.59AEE948 pKa = 4.22KK949 pKa = 10.83LHH951 pKa = 6.51MYY953 pKa = 9.61SASLIGGMVLGGFTSAAALPFSYY976 pKa = 10.43AVQARR981 pKa = 11.84LNYY984 pKa = 10.03LALQTDD990 pKa = 4.07VLQRR994 pKa = 11.84NQQLLAEE1001 pKa = 4.79SFNSAIGNITSAFEE1015 pKa = 4.23SVKK1018 pKa = 10.42EE1019 pKa = 4.47AISQTSKK1026 pKa = 11.06GLNTVAHH1033 pKa = 7.25ALTKK1037 pKa = 9.6VQEE1040 pKa = 4.43VVNSQGAALTQLTVQLQHH1058 pKa = 6.2NFQAISSSIDD1068 pKa = 3.66DD1069 pKa = 3.63IYY1071 pKa = 11.74SRR1073 pKa = 11.84LDD1075 pKa = 3.15ILSADD1080 pKa = 3.79VQVDD1084 pKa = 3.36RR1085 pKa = 11.84LITGRR1090 pKa = 11.84LSALNAFVSQTLTKK1104 pKa = 8.56YY1105 pKa = 9.43TEE1107 pKa = 4.15VQASRR1112 pKa = 11.84KK1113 pKa = 9.06LAQQKK1118 pKa = 10.1VNEE1121 pKa = 4.45CVKK1124 pKa = 10.67SQSQRR1129 pKa = 11.84YY1130 pKa = 7.48GFCGGDD1136 pKa = 3.54GEE1138 pKa = 5.79HH1139 pKa = 6.85IFSLVQAAPQGLLFLHH1155 pKa = 6.22TVLVPSDD1162 pKa = 3.98FVDD1165 pKa = 4.92VIAIAGLCVNDD1176 pKa = 5.21EE1177 pKa = 3.94IALTLRR1183 pKa = 11.84EE1184 pKa = 4.29PGLVLFTHH1192 pKa = 6.92EE1193 pKa = 4.72LQNHH1197 pKa = 5.41TATEE1201 pKa = 4.39YY1202 pKa = 10.01FVSSRR1207 pKa = 11.84RR1208 pKa = 11.84MFEE1211 pKa = 3.62PRR1213 pKa = 11.84KK1214 pKa = 8.1PTVSDD1219 pKa = 4.03FVQIEE1224 pKa = 4.69SCVVTYY1230 pKa = 11.26VNLTRR1235 pKa = 11.84DD1236 pKa = 3.63QLPDD1240 pKa = 3.9VIPDD1244 pKa = 3.77YY1245 pKa = 11.31IDD1247 pKa = 3.25VNKK1250 pKa = 9.69TLDD1253 pKa = 4.12EE1254 pKa = 4.36ILASLPNRR1262 pKa = 11.84TGPSLPLDD1270 pKa = 3.87VFNATYY1276 pKa = 11.15LNLTGEE1282 pKa = 4.35IADD1285 pKa = 3.79LEE1287 pKa = 4.28QRR1289 pKa = 11.84SEE1291 pKa = 4.11SLRR1294 pKa = 11.84NTTEE1298 pKa = 3.87EE1299 pKa = 4.15LQSLIYY1305 pKa = 10.39NINNTLVDD1313 pKa = 4.54LEE1315 pKa = 4.17WLNRR1319 pKa = 11.84VEE1321 pKa = 5.97TYY1323 pKa = 10.41IKK1325 pKa = 9.23WPWWVWLIIFIVLIFVVSLLVFCCISTGCCGCCGCCCACFSGCCRR1370 pKa = 11.84GPRR1373 pKa = 11.84LQPYY1377 pKa = 8.78EE1378 pKa = 4.19VFEE1381 pKa = 4.36KK1382 pKa = 11.03VHH1384 pKa = 5.02VQQ1386 pKa = 3.02
MM1 pKa = 7.99KK2 pKa = 10.42SLTYY6 pKa = 9.83FWLFLPVLSTLSLPQDD22 pKa = 3.57VTRR25 pKa = 11.84CSANTNFRR33 pKa = 11.84RR34 pKa = 11.84FFSKK38 pKa = 10.72FNVQAPAVVVLGGYY52 pKa = 10.04LPIGEE57 pKa = 4.23NQGVNSTWYY66 pKa = 10.45CAGQHH71 pKa = 5.25PTASGVHH78 pKa = 6.7GIFVSHH84 pKa = 6.55IRR86 pKa = 11.84GGHH89 pKa = 4.59GFEE92 pKa = 4.58IGISQEE98 pKa = 3.85PFDD101 pKa = 4.04PSGYY105 pKa = 9.89QLYY108 pKa = 10.0LHH110 pKa = 7.06KK111 pKa = 10.35ATNGNTNATARR122 pKa = 11.84LRR124 pKa = 11.84ICQFPSIKK132 pKa = 9.87TLGPTANNDD141 pKa = 2.87VTTGRR146 pKa = 11.84NCLFNKK152 pKa = 9.98AIPAHH157 pKa = 5.62MSEE160 pKa = 4.46HH161 pKa = 5.85SVVGITWDD169 pKa = 3.48NDD171 pKa = 3.08RR172 pKa = 11.84VTVFSDD178 pKa = 3.69KK179 pKa = 10.46IYY181 pKa = 11.02YY182 pKa = 10.22FYY184 pKa = 10.98FKK186 pKa = 10.95NDD188 pKa = 3.08WSRR191 pKa = 11.84VATKK195 pKa = 10.01CYY197 pKa = 10.67NSGGCAMQYY206 pKa = 10.42VYY208 pKa = 11.04EE209 pKa = 4.08PTYY212 pKa = 11.64YY213 pKa = 9.87MLNVTSAGEE222 pKa = 4.14DD223 pKa = 3.74GISYY227 pKa = 9.02QPCTANCIGYY237 pKa = 9.17AANVFATEE245 pKa = 4.27PNGHH249 pKa = 6.21IPEE252 pKa = 4.55GFSFNNWFLLSNDD265 pKa = 3.05STLVHH270 pKa = 6.24GKK272 pKa = 9.44VVSNQPLLVNCLLAIPKK289 pKa = 9.74IYY291 pKa = 10.87GLGQFFSFNQTIDD304 pKa = 3.8GVCNGAAVQRR314 pKa = 11.84APEE317 pKa = 3.93ALRR320 pKa = 11.84FNINDD325 pKa = 3.42TSVILAEE332 pKa = 4.36GSIVLHH338 pKa = 6.0TALGTNFSFVCSNSSNPHH356 pKa = 6.09LATFAIPLGATQVPYY371 pKa = 11.25YY372 pKa = 10.63CFLKK376 pKa = 10.57VDD378 pKa = 4.51TYY380 pKa = 11.79NSTVYY385 pKa = 10.65KK386 pKa = 9.87FLAVLPPTVRR396 pKa = 11.84EE397 pKa = 3.97IVITKK402 pKa = 10.54YY403 pKa = 10.93GDD405 pKa = 3.33VYY407 pKa = 11.7VNGFGYY413 pKa = 10.69LHH415 pKa = 7.17LGLLDD420 pKa = 3.63AVTINFTGHH429 pKa = 5.73GTDD432 pKa = 4.59DD433 pKa = 4.08DD434 pKa = 5.06VSGFWTIASTNFVDD448 pKa = 5.9ALIEE452 pKa = 4.16VQGTAIQRR460 pKa = 11.84ILYY463 pKa = 10.06CDD465 pKa = 4.15DD466 pKa = 4.14PVSQLKK472 pKa = 10.32CSQVAFDD479 pKa = 5.4LDD481 pKa = 3.89DD482 pKa = 3.81GFYY485 pKa = 10.38PISSRR490 pKa = 11.84NLLSHH495 pKa = 6.55EE496 pKa = 4.12QPISFVTLPSFNDD509 pKa = 3.26HH510 pKa = 6.27SFVNITVSASFGGHH524 pKa = 5.66SGANLIASDD533 pKa = 3.51TTINGFSSFCVDD545 pKa = 2.78TRR547 pKa = 11.84QFTISLFYY555 pKa = 11.08NVTNSYY561 pKa = 11.02GYY563 pKa = 10.2VSKK566 pKa = 10.92SQDD569 pKa = 3.46SNCPFTLQSVNDD581 pKa = 3.71YY582 pKa = 11.36LSFSKK587 pKa = 10.79FCVSTSLLASACTIDD602 pKa = 4.13LFGYY606 pKa = 9.44PEE608 pKa = 4.41FGSGVKK614 pKa = 8.38FTSLYY619 pKa = 10.0FQFTKK624 pKa = 11.13GEE626 pKa = 4.85LITGTPKK633 pKa = 10.48PLEE636 pKa = 4.31GVTDD640 pKa = 3.65VSFMTLDD647 pKa = 3.0VCTKK651 pKa = 7.34YY652 pKa = 9.42TIYY655 pKa = 10.66GFKK658 pKa = 10.88GEE660 pKa = 4.78GIITLTNSSFLAGVYY675 pKa = 7.31YY676 pKa = 10.55TSDD679 pKa = 3.25SGQLLAFKK687 pKa = 10.21NVTSGAVYY695 pKa = 10.52SVTPCSFSEE704 pKa = 3.9QAAYY708 pKa = 10.85VDD710 pKa = 4.13DD711 pKa = 6.26DD712 pKa = 3.81IVGVISSLSSSTFNSTRR729 pKa = 11.84EE730 pKa = 3.91LPGFFYY736 pKa = 10.78HH737 pKa = 7.56SNDD740 pKa = 2.92GSNCTEE746 pKa = 3.82PVLVYY751 pKa = 11.07SNIGVCKK758 pKa = 9.81SGSIGYY764 pKa = 9.24VPSQSGQVKK773 pKa = 9.94IAPTVTGNISIPTNFSMSIRR793 pKa = 11.84TEE795 pKa = 3.69YY796 pKa = 10.03LQLYY800 pKa = 6.15NTPVSVDD807 pKa = 3.13CATYY811 pKa = 9.95VCNGNSRR818 pKa = 11.84CKK820 pKa = 10.28QLLTQYY826 pKa = 9.3TAACKK831 pKa = 9.6TIEE834 pKa = 4.39SALQLSARR842 pKa = 11.84LEE844 pKa = 4.32SVEE847 pKa = 4.22VNSMLTISEE856 pKa = 4.32EE857 pKa = 4.08ALQLATISSFNGDD870 pKa = 3.29GYY872 pKa = 11.6NFTNVLGVSVYY883 pKa = 10.82DD884 pKa = 3.71PASGRR889 pKa = 11.84VVQKK893 pKa = 10.67RR894 pKa = 11.84SFIEE898 pKa = 4.23DD899 pKa = 3.54LLFNKK904 pKa = 9.39VVTNGLGTVDD914 pKa = 4.28EE915 pKa = 4.8DD916 pKa = 4.8YY917 pKa = 11.12KK918 pKa = 10.79RR919 pKa = 11.84CSNGRR924 pKa = 11.84SVADD928 pKa = 3.65LVCAQYY934 pKa = 11.29YY935 pKa = 9.11SGVMVLPGVVDD946 pKa = 3.59AEE948 pKa = 4.22KK949 pKa = 10.83LHH951 pKa = 6.51MYY953 pKa = 9.61SASLIGGMVLGGFTSAAALPFSYY976 pKa = 10.43AVQARR981 pKa = 11.84LNYY984 pKa = 10.03LALQTDD990 pKa = 4.07VLQRR994 pKa = 11.84NQQLLAEE1001 pKa = 4.79SFNSAIGNITSAFEE1015 pKa = 4.23SVKK1018 pKa = 10.42EE1019 pKa = 4.47AISQTSKK1026 pKa = 11.06GLNTVAHH1033 pKa = 7.25ALTKK1037 pKa = 9.6VQEE1040 pKa = 4.43VVNSQGAALTQLTVQLQHH1058 pKa = 6.2NFQAISSSIDD1068 pKa = 3.66DD1069 pKa = 3.63IYY1071 pKa = 11.74SRR1073 pKa = 11.84LDD1075 pKa = 3.15ILSADD1080 pKa = 3.79VQVDD1084 pKa = 3.36RR1085 pKa = 11.84LITGRR1090 pKa = 11.84LSALNAFVSQTLTKK1104 pKa = 8.56YY1105 pKa = 9.43TEE1107 pKa = 4.15VQASRR1112 pKa = 11.84KK1113 pKa = 9.06LAQQKK1118 pKa = 10.1VNEE1121 pKa = 4.45CVKK1124 pKa = 10.67SQSQRR1129 pKa = 11.84YY1130 pKa = 7.48GFCGGDD1136 pKa = 3.54GEE1138 pKa = 5.79HH1139 pKa = 6.85IFSLVQAAPQGLLFLHH1155 pKa = 6.22TVLVPSDD1162 pKa = 3.98FVDD1165 pKa = 4.92VIAIAGLCVNDD1176 pKa = 5.21EE1177 pKa = 3.94IALTLRR1183 pKa = 11.84EE1184 pKa = 4.29PGLVLFTHH1192 pKa = 6.92EE1193 pKa = 4.72LQNHH1197 pKa = 5.41TATEE1201 pKa = 4.39YY1202 pKa = 10.01FVSSRR1207 pKa = 11.84RR1208 pKa = 11.84MFEE1211 pKa = 3.62PRR1213 pKa = 11.84KK1214 pKa = 8.1PTVSDD1219 pKa = 4.03FVQIEE1224 pKa = 4.69SCVVTYY1230 pKa = 11.26VNLTRR1235 pKa = 11.84DD1236 pKa = 3.63QLPDD1240 pKa = 3.9VIPDD1244 pKa = 3.77YY1245 pKa = 11.31IDD1247 pKa = 3.25VNKK1250 pKa = 9.69TLDD1253 pKa = 4.12EE1254 pKa = 4.36ILASLPNRR1262 pKa = 11.84TGPSLPLDD1270 pKa = 3.87VFNATYY1276 pKa = 11.15LNLTGEE1282 pKa = 4.35IADD1285 pKa = 3.79LEE1287 pKa = 4.28QRR1289 pKa = 11.84SEE1291 pKa = 4.11SLRR1294 pKa = 11.84NTTEE1298 pKa = 3.87EE1299 pKa = 4.15LQSLIYY1305 pKa = 10.39NINNTLVDD1313 pKa = 4.54LEE1315 pKa = 4.17WLNRR1319 pKa = 11.84VEE1321 pKa = 5.97TYY1323 pKa = 10.41IKK1325 pKa = 9.23WPWWVWLIIFIVLIFVVSLLVFCCISTGCCGCCGCCCACFSGCCRR1370 pKa = 11.84GPRR1373 pKa = 11.84LQPYY1377 pKa = 8.78EE1378 pKa = 4.19VFEE1381 pKa = 4.36KK1382 pKa = 11.03VHH1384 pKa = 5.02VQQ1386 pKa = 3.02
Molecular weight: 151.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H6WQB8|H6WQB8_9ALPC Envelope small membrane protein OS=Porcine epidemic diarrhea virus OX=28295 GN=E PE=3 SV=1
MM1 pKa = 7.63ASVSFQDD8 pKa = 3.2RR9 pKa = 11.84GRR11 pKa = 11.84KK12 pKa = 8.69RR13 pKa = 11.84VPLSLYY19 pKa = 10.66APLRR23 pKa = 11.84VTNDD27 pKa = 2.99KK28 pKa = 10.52PLSKK32 pKa = 10.59VLANNAVPTNKK43 pKa = 9.77GNKK46 pKa = 8.6DD47 pKa = 3.38QQIGYY52 pKa = 9.0WNEE55 pKa = 3.64QIRR58 pKa = 11.84WRR60 pKa = 11.84MRR62 pKa = 11.84RR63 pKa = 11.84GEE65 pKa = 4.48RR66 pKa = 11.84IEE68 pKa = 4.86QPSNWHH74 pKa = 6.49FYY76 pKa = 10.41YY77 pKa = 10.82LGTGPHH83 pKa = 6.47ADD85 pKa = 2.9LRR87 pKa = 11.84YY88 pKa = 7.7RR89 pKa = 11.84TRR91 pKa = 11.84TEE93 pKa = 3.79GVFWVAKK100 pKa = 9.55EE101 pKa = 4.01GAKK104 pKa = 9.42TEE106 pKa = 4.04PTNLGVRR113 pKa = 11.84KK114 pKa = 9.87ASEE117 pKa = 4.02KK118 pKa = 10.49PIIPNFSQQLPSVVEE133 pKa = 3.81IVEE136 pKa = 4.45PNTPPTSRR144 pKa = 11.84ANSRR148 pKa = 11.84SRR150 pKa = 11.84SRR152 pKa = 11.84GNGNNRR158 pKa = 11.84SRR160 pKa = 11.84SPSNNRR166 pKa = 11.84GNNQSRR172 pKa = 11.84GNSQNRR178 pKa = 11.84GNNQGRR184 pKa = 11.84GASQNRR190 pKa = 11.84GGNNNNNNKK199 pKa = 9.52SRR201 pKa = 11.84NQSKK205 pKa = 10.03NRR207 pKa = 11.84NQSNDD212 pKa = 3.15RR213 pKa = 11.84GGVTSRR219 pKa = 11.84DD220 pKa = 3.65DD221 pKa = 3.36LVAAVKK227 pKa = 10.47DD228 pKa = 3.79ALKK231 pKa = 10.92SLGIGEE237 pKa = 4.79NPDD240 pKa = 4.07KK241 pKa = 11.01LKK243 pKa = 10.46QQQKK247 pKa = 9.45PKK249 pKa = 10.0QEE251 pKa = 4.27RR252 pKa = 11.84SDD254 pKa = 3.7SSGKK258 pKa = 8.11NTPKK262 pKa = 10.27KK263 pKa = 9.59NKK265 pKa = 9.58SRR267 pKa = 11.84ATSKK271 pKa = 10.69EE272 pKa = 3.65RR273 pKa = 11.84DD274 pKa = 3.56LKK276 pKa = 10.98DD277 pKa = 3.0IPEE280 pKa = 3.86WRR282 pKa = 11.84RR283 pKa = 11.84IPKK286 pKa = 10.29GEE288 pKa = 4.08NSVAACFGPRR298 pKa = 11.84GGFKK302 pKa = 10.83NFGDD306 pKa = 3.81AEE308 pKa = 4.15FVEE311 pKa = 4.62KK312 pKa = 10.79GVDD315 pKa = 3.14ASGYY319 pKa = 9.79AQIASLAPNVAALLFGGNVAVRR341 pKa = 11.84EE342 pKa = 4.12LADD345 pKa = 3.7SYY347 pKa = 11.34EE348 pKa = 3.54ITYY351 pKa = 9.37NYY353 pKa = 10.28KK354 pKa = 8.23MTVPKK359 pKa = 10.33SDD361 pKa = 4.0PNVEE365 pKa = 4.18LLVSQVDD372 pKa = 3.54AFKK375 pKa = 10.5TGNAKK380 pKa = 9.27PQRR383 pKa = 11.84KK384 pKa = 8.83KK385 pKa = 9.71EE386 pKa = 4.06KK387 pKa = 9.23KK388 pKa = 9.21NKK390 pKa = 10.14RR391 pKa = 11.84EE392 pKa = 3.78TTQQLNEE399 pKa = 3.73EE400 pKa = 4.81AIYY403 pKa = 11.05DD404 pKa = 3.99DD405 pKa = 3.93VGVPSDD411 pKa = 3.56VTHH414 pKa = 7.56ANLEE418 pKa = 3.75WDD420 pKa = 3.83TAVDD424 pKa = 3.73GGDD427 pKa = 3.32TAVEE431 pKa = 4.31IINEE435 pKa = 4.01IFDD438 pKa = 3.69TGNN441 pKa = 2.86
MM1 pKa = 7.63ASVSFQDD8 pKa = 3.2RR9 pKa = 11.84GRR11 pKa = 11.84KK12 pKa = 8.69RR13 pKa = 11.84VPLSLYY19 pKa = 10.66APLRR23 pKa = 11.84VTNDD27 pKa = 2.99KK28 pKa = 10.52PLSKK32 pKa = 10.59VLANNAVPTNKK43 pKa = 9.77GNKK46 pKa = 8.6DD47 pKa = 3.38QQIGYY52 pKa = 9.0WNEE55 pKa = 3.64QIRR58 pKa = 11.84WRR60 pKa = 11.84MRR62 pKa = 11.84RR63 pKa = 11.84GEE65 pKa = 4.48RR66 pKa = 11.84IEE68 pKa = 4.86QPSNWHH74 pKa = 6.49FYY76 pKa = 10.41YY77 pKa = 10.82LGTGPHH83 pKa = 6.47ADD85 pKa = 2.9LRR87 pKa = 11.84YY88 pKa = 7.7RR89 pKa = 11.84TRR91 pKa = 11.84TEE93 pKa = 3.79GVFWVAKK100 pKa = 9.55EE101 pKa = 4.01GAKK104 pKa = 9.42TEE106 pKa = 4.04PTNLGVRR113 pKa = 11.84KK114 pKa = 9.87ASEE117 pKa = 4.02KK118 pKa = 10.49PIIPNFSQQLPSVVEE133 pKa = 3.81IVEE136 pKa = 4.45PNTPPTSRR144 pKa = 11.84ANSRR148 pKa = 11.84SRR150 pKa = 11.84SRR152 pKa = 11.84GNGNNRR158 pKa = 11.84SRR160 pKa = 11.84SPSNNRR166 pKa = 11.84GNNQSRR172 pKa = 11.84GNSQNRR178 pKa = 11.84GNNQGRR184 pKa = 11.84GASQNRR190 pKa = 11.84GGNNNNNNKK199 pKa = 9.52SRR201 pKa = 11.84NQSKK205 pKa = 10.03NRR207 pKa = 11.84NQSNDD212 pKa = 3.15RR213 pKa = 11.84GGVTSRR219 pKa = 11.84DD220 pKa = 3.65DD221 pKa = 3.36LVAAVKK227 pKa = 10.47DD228 pKa = 3.79ALKK231 pKa = 10.92SLGIGEE237 pKa = 4.79NPDD240 pKa = 4.07KK241 pKa = 11.01LKK243 pKa = 10.46QQQKK247 pKa = 9.45PKK249 pKa = 10.0QEE251 pKa = 4.27RR252 pKa = 11.84SDD254 pKa = 3.7SSGKK258 pKa = 8.11NTPKK262 pKa = 10.27KK263 pKa = 9.59NKK265 pKa = 9.58SRR267 pKa = 11.84ATSKK271 pKa = 10.69EE272 pKa = 3.65RR273 pKa = 11.84DD274 pKa = 3.56LKK276 pKa = 10.98DD277 pKa = 3.0IPEE280 pKa = 3.86WRR282 pKa = 11.84RR283 pKa = 11.84IPKK286 pKa = 10.29GEE288 pKa = 4.08NSVAACFGPRR298 pKa = 11.84GGFKK302 pKa = 10.83NFGDD306 pKa = 3.81AEE308 pKa = 4.15FVEE311 pKa = 4.62KK312 pKa = 10.79GVDD315 pKa = 3.14ASGYY319 pKa = 9.79AQIASLAPNVAALLFGGNVAVRR341 pKa = 11.84EE342 pKa = 4.12LADD345 pKa = 3.7SYY347 pKa = 11.34EE348 pKa = 3.54ITYY351 pKa = 9.37NYY353 pKa = 10.28KK354 pKa = 8.23MTVPKK359 pKa = 10.33SDD361 pKa = 4.0PNVEE365 pKa = 4.18LLVSQVDD372 pKa = 3.54AFKK375 pKa = 10.5TGNAKK380 pKa = 9.27PQRR383 pKa = 11.84KK384 pKa = 8.83KK385 pKa = 9.71EE386 pKa = 4.06KK387 pKa = 9.23KK388 pKa = 9.21NKK390 pKa = 10.14RR391 pKa = 11.84EE392 pKa = 3.78TTQQLNEE399 pKa = 3.73EE400 pKa = 4.81AIYY403 pKa = 11.05DD404 pKa = 3.99DD405 pKa = 3.93VGVPSDD411 pKa = 3.56VTHH414 pKa = 7.56ANLEE418 pKa = 3.75WDD420 pKa = 3.83TAVDD424 pKa = 3.73GGDD427 pKa = 3.32TAVEE431 pKa = 4.31IINEE435 pKa = 4.01IFDD438 pKa = 3.69TGNN441 pKa = 2.86
Molecular weight: 48.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6302 |
76 |
3974 |
1050.3 |
115.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.061 ± 0.393 | 2.92 ± 0.603 |
5.173 ± 0.437 | 3.951 ± 0.346 |
6.062 ± 0.674 | 7.061 ± 0.304 |
1.698 ± 0.233 | 4.967 ± 0.477 |
5.03 ± 0.813 | 9.092 ± 0.759 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.619 ± 0.362 | 5.522 ± 0.932 |
3.586 ± 0.366 | 3.078 ± 0.708 |
3.396 ± 0.977 | 7.839 ± 0.651 |
6.331 ± 0.487 | 10.235 ± 0.871 |
1.127 ± 0.339 | 4.253 ± 0.32 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
