
Gyrovirus 4
Taxonomy: Viruses; Anelloviridae; Gyrovirus; Gyrovirus homsa3
Average proteome isoelectric point is 7.52
Get precalculated fractions of proteins
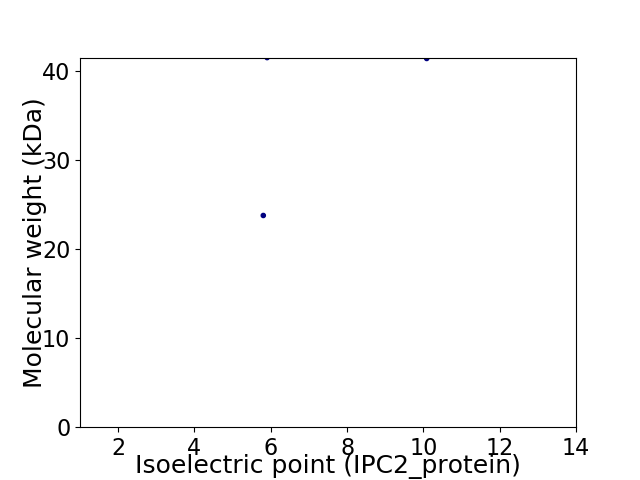
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I7DCG0|I7DCG0_9VIRU CA1 OS=Gyrovirus 4 OX=1214955 GN=VP1 PE=3 SV=1
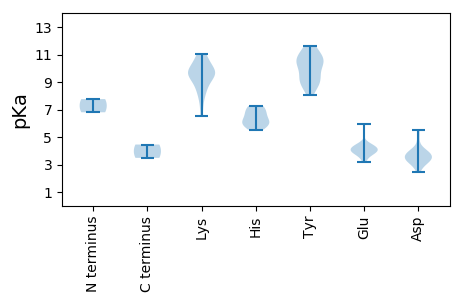
MM1 pKa = 7.76GSPDD5 pKa = 3.32HH6 pKa = 6.91RR7 pKa = 11.84FSAVPIDD14 pKa = 5.03LLDD17 pKa = 3.95SVCPQWKK24 pKa = 8.77QVCFNYY30 pKa = 10.66GIACWLTEE38 pKa = 3.75CRR40 pKa = 11.84VSHH43 pKa = 6.65SRR45 pKa = 11.84VCRR48 pKa = 11.84CTSFRR53 pKa = 11.84NHH55 pKa = 5.54WFQLEE60 pKa = 4.2GKK62 pKa = 8.42PVSTVEE68 pKa = 4.31VGVQTDD74 pKa = 3.88PGDD77 pKa = 3.91LEE79 pKa = 4.39PPRR82 pKa = 11.84SGTSTAEE89 pKa = 3.2IGPRR93 pKa = 11.84VDD95 pKa = 2.8AARR98 pKa = 11.84RR99 pKa = 11.84LLSSISFKK107 pKa = 10.72RR108 pKa = 11.84DD109 pKa = 2.47ACRR112 pKa = 11.84ALEE115 pKa = 4.21GAPKK119 pKa = 9.63PKK121 pKa = 9.68KK122 pKa = 9.8RR123 pKa = 11.84KK124 pKa = 9.85RR125 pKa = 11.84GDD127 pKa = 3.02ARR129 pKa = 11.84KK130 pKa = 9.49EE131 pKa = 3.36FLRR134 pKa = 11.84RR135 pKa = 11.84FRR137 pKa = 11.84EE138 pKa = 4.52GEE140 pKa = 3.98EE141 pKa = 4.29TEE143 pKa = 5.94SDD145 pKa = 3.98AEE147 pKa = 3.88WDD149 pKa = 4.37SFVEE153 pKa = 4.34WSDD156 pKa = 3.42SDD158 pKa = 3.66GDD160 pKa = 3.94PGVDD164 pKa = 3.57YY165 pKa = 10.32MRR167 pKa = 11.84GGAGGGILRR176 pKa = 11.84EE177 pKa = 4.07DD178 pKa = 3.47RR179 pKa = 11.84MGGGIEE185 pKa = 4.06VEE187 pKa = 4.38STGGGTGGTEE197 pKa = 3.65GTPFLKK203 pKa = 10.65LLSGNFTSSTPARR216 pKa = 11.84MM217 pKa = 3.49
MM1 pKa = 7.76GSPDD5 pKa = 3.32HH6 pKa = 6.91RR7 pKa = 11.84FSAVPIDD14 pKa = 5.03LLDD17 pKa = 3.95SVCPQWKK24 pKa = 8.77QVCFNYY30 pKa = 10.66GIACWLTEE38 pKa = 3.75CRR40 pKa = 11.84VSHH43 pKa = 6.65SRR45 pKa = 11.84VCRR48 pKa = 11.84CTSFRR53 pKa = 11.84NHH55 pKa = 5.54WFQLEE60 pKa = 4.2GKK62 pKa = 8.42PVSTVEE68 pKa = 4.31VGVQTDD74 pKa = 3.88PGDD77 pKa = 3.91LEE79 pKa = 4.39PPRR82 pKa = 11.84SGTSTAEE89 pKa = 3.2IGPRR93 pKa = 11.84VDD95 pKa = 2.8AARR98 pKa = 11.84RR99 pKa = 11.84LLSSISFKK107 pKa = 10.72RR108 pKa = 11.84DD109 pKa = 2.47ACRR112 pKa = 11.84ALEE115 pKa = 4.21GAPKK119 pKa = 9.63PKK121 pKa = 9.68KK122 pKa = 9.8RR123 pKa = 11.84KK124 pKa = 9.85RR125 pKa = 11.84GDD127 pKa = 3.02ARR129 pKa = 11.84KK130 pKa = 9.49EE131 pKa = 3.36FLRR134 pKa = 11.84RR135 pKa = 11.84FRR137 pKa = 11.84EE138 pKa = 4.52GEE140 pKa = 3.98EE141 pKa = 4.29TEE143 pKa = 5.94SDD145 pKa = 3.98AEE147 pKa = 3.88WDD149 pKa = 4.37SFVEE153 pKa = 4.34WSDD156 pKa = 3.42SDD158 pKa = 3.66GDD160 pKa = 3.94PGVDD164 pKa = 3.57YY165 pKa = 10.32MRR167 pKa = 11.84GGAGGGILRR176 pKa = 11.84EE177 pKa = 4.07DD178 pKa = 3.47RR179 pKa = 11.84MGGGIEE185 pKa = 4.06VEE187 pKa = 4.38STGGGTGGTEE197 pKa = 3.65GTPFLKK203 pKa = 10.65LLSGNFTSSTPARR216 pKa = 11.84MM217 pKa = 3.49
Molecular weight: 23.77 kDa
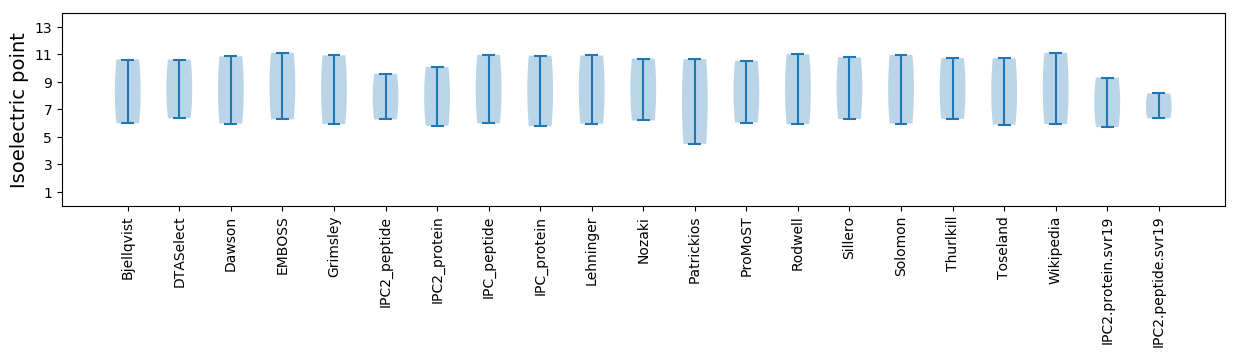
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I7DCG0|I7DCG0_9VIRU CA1 OS=Gyrovirus 4 OX=1214955 GN=VP1 PE=3 SV=1
MM1 pKa = 6.8VRR3 pKa = 11.84FRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84PWGRR11 pKa = 11.84LYY13 pKa = 10.84AWRR16 pKa = 11.84RR17 pKa = 11.84GGWHH21 pKa = 6.77FKK23 pKa = 9.76RR24 pKa = 11.84RR25 pKa = 11.84PYY27 pKa = 9.81GRR29 pKa = 11.84WYY31 pKa = 10.62RR32 pKa = 11.84SGKK35 pKa = 7.56YY36 pKa = 8.05RR37 pKa = 11.84RR38 pKa = 11.84GYY40 pKa = 9.25RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 5.78RR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 10.62AFSKK50 pKa = 8.59AFKK53 pKa = 9.67RR54 pKa = 11.84KK55 pKa = 9.52FYY57 pKa = 9.72IQHH60 pKa = 6.33PGSYY64 pKa = 9.24VVRR67 pKa = 11.84KK68 pKa = 6.55TRR70 pKa = 11.84PYY72 pKa = 10.54NSMKK76 pKa = 10.38VFFQGVIFVNNAVFQSGQAEE96 pKa = 3.95TSAFRR101 pKa = 11.84VYY103 pKa = 10.55SIEE106 pKa = 3.98LSLRR110 pKa = 11.84NLLLAYY116 pKa = 9.15YY117 pKa = 8.72VQYY120 pKa = 11.36VSGGPGWTGTSAAKK134 pKa = 10.78VNISPLDD141 pKa = 3.29WWRR144 pKa = 11.84WAYY147 pKa = 10.1IRR149 pKa = 11.84MHH151 pKa = 7.26PSPDD155 pKa = 2.98QRR157 pKa = 11.84VFANATRR164 pKa = 11.84PTDD167 pKa = 3.56TQLFNYY173 pKa = 8.88FSQWQLFRR181 pKa = 11.84HH182 pKa = 6.16IKK184 pKa = 9.02TDD186 pKa = 3.9FRR188 pKa = 11.84VLNTDD193 pKa = 3.19VTNTPDD199 pKa = 3.39VRR201 pKa = 11.84ILASLLTQDD210 pKa = 5.54SYY212 pKa = 11.63WQVQNKK218 pKa = 8.12TSADD222 pKa = 3.36LSQLKK227 pKa = 9.99FPSFAEE233 pKa = 3.99LTAWGAQWAFLPGQKK248 pKa = 9.72SVSGRR253 pKa = 11.84SFNHH257 pKa = 5.86HH258 pKa = 5.96SMRR261 pKa = 11.84GTNDD265 pKa = 3.26PKK267 pKa = 11.06GEE269 pKa = 3.98PWKK272 pKa = 9.49TLTPLQHH279 pKa = 6.18QVVSDD284 pKa = 3.7INWVFGTLFLAEE296 pKa = 4.46FSNFTVLNEE305 pKa = 3.95YY306 pKa = 8.8NTEE309 pKa = 3.75NRR311 pKa = 11.84ALAATGLSEE320 pKa = 3.7TWSIVKK326 pKa = 9.48VRR328 pKa = 11.84SLFEE332 pKa = 4.18FGNQQYY338 pKa = 11.3LLADD342 pKa = 3.65SVRR345 pKa = 11.84FTSHH349 pKa = 7.21LPTT352 pKa = 4.46
MM1 pKa = 6.8VRR3 pKa = 11.84FRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84PWGRR11 pKa = 11.84LYY13 pKa = 10.84AWRR16 pKa = 11.84RR17 pKa = 11.84GGWHH21 pKa = 6.77FKK23 pKa = 9.76RR24 pKa = 11.84RR25 pKa = 11.84PYY27 pKa = 9.81GRR29 pKa = 11.84WYY31 pKa = 10.62RR32 pKa = 11.84SGKK35 pKa = 7.56YY36 pKa = 8.05RR37 pKa = 11.84RR38 pKa = 11.84GYY40 pKa = 9.25RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 5.78RR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 10.62AFSKK50 pKa = 8.59AFKK53 pKa = 9.67RR54 pKa = 11.84KK55 pKa = 9.52FYY57 pKa = 9.72IQHH60 pKa = 6.33PGSYY64 pKa = 9.24VVRR67 pKa = 11.84KK68 pKa = 6.55TRR70 pKa = 11.84PYY72 pKa = 10.54NSMKK76 pKa = 10.38VFFQGVIFVNNAVFQSGQAEE96 pKa = 3.95TSAFRR101 pKa = 11.84VYY103 pKa = 10.55SIEE106 pKa = 3.98LSLRR110 pKa = 11.84NLLLAYY116 pKa = 9.15YY117 pKa = 8.72VQYY120 pKa = 11.36VSGGPGWTGTSAAKK134 pKa = 10.78VNISPLDD141 pKa = 3.29WWRR144 pKa = 11.84WAYY147 pKa = 10.1IRR149 pKa = 11.84MHH151 pKa = 7.26PSPDD155 pKa = 2.98QRR157 pKa = 11.84VFANATRR164 pKa = 11.84PTDD167 pKa = 3.56TQLFNYY173 pKa = 8.88FSQWQLFRR181 pKa = 11.84HH182 pKa = 6.16IKK184 pKa = 9.02TDD186 pKa = 3.9FRR188 pKa = 11.84VLNTDD193 pKa = 3.19VTNTPDD199 pKa = 3.39VRR201 pKa = 11.84ILASLLTQDD210 pKa = 5.54SYY212 pKa = 11.63WQVQNKK218 pKa = 8.12TSADD222 pKa = 3.36LSQLKK227 pKa = 9.99FPSFAEE233 pKa = 3.99LTAWGAQWAFLPGQKK248 pKa = 9.72SVSGRR253 pKa = 11.84SFNHH257 pKa = 5.86HH258 pKa = 5.96SMRR261 pKa = 11.84GTNDD265 pKa = 3.26PKK267 pKa = 11.06GEE269 pKa = 3.98PWKK272 pKa = 9.49TLTPLQHH279 pKa = 6.18QVVSDD284 pKa = 3.7INWVFGTLFLAEE296 pKa = 4.46FSNFTVLNEE305 pKa = 3.95YY306 pKa = 8.8NTEE309 pKa = 3.75NRR311 pKa = 11.84ALAATGLSEE320 pKa = 3.7TWSIVKK326 pKa = 9.48VRR328 pKa = 11.84SLFEE332 pKa = 4.18FGNQQYY338 pKa = 11.3LLADD342 pKa = 3.65SVRR345 pKa = 11.84FTSHH349 pKa = 7.21LPTT352 pKa = 4.46
Molecular weight: 41.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
569 |
217 |
352 |
284.5 |
32.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.151 ± 0.39 | 1.23 ± 1.252 |
4.569 ± 1.469 | 4.569 ± 2.047 |
6.327 ± 1.078 | 8.436 ± 2.513 |
2.109 ± 0.456 | 2.636 ± 0.081 |
4.218 ± 0.044 | 7.206 ± 0.762 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.406 ± 0.274 | 3.691 ± 1.448 |
5.097 ± 0.561 | 4.042 ± 1.379 |
9.666 ± 0.007 | 8.612 ± 0.379 |
6.503 ± 0.321 | 6.503 ± 0.321 |
3.515 ± 0.759 | 3.515 ± 1.626 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
