
Hubei orthoptera virus 5
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Xinmoviridae; Anphevirus; Orthopteran anphevirus
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
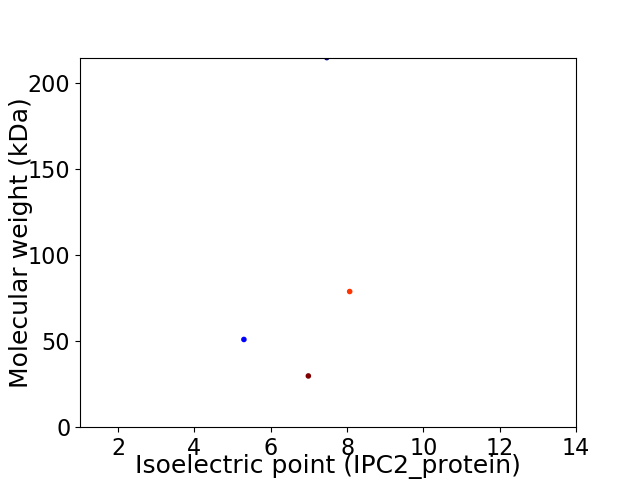
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KNE4|A0A1L3KNE4_9MONO Uncharacterized protein OS=Hubei orthoptera virus 5 OX=1923013 PE=4 SV=1
MM1 pKa = 7.92DD2 pKa = 4.8TFQIDD7 pKa = 3.76PASDD11 pKa = 3.71TPCPHH16 pKa = 6.92RR17 pKa = 11.84AIVFQPAGGRR27 pKa = 11.84SVRR30 pKa = 11.84IYY32 pKa = 10.97ARR34 pKa = 11.84PPDD37 pKa = 3.66QGAAPSYY44 pKa = 10.14PIYY47 pKa = 10.55LCPNADD53 pKa = 3.82FLTLSVDD60 pKa = 3.03ALTIGGYY67 pKa = 9.99GDD69 pKa = 3.56HH70 pKa = 6.21VQYY73 pKa = 10.81DD74 pKa = 3.51IRR76 pKa = 11.84NQIEE80 pKa = 4.0PQDD83 pKa = 3.77WVADD87 pKa = 3.75SATTFTVAPSRR98 pKa = 11.84LPVWIGIDD106 pKa = 3.29HH107 pKa = 7.17AAAKK111 pKa = 10.11YY112 pKa = 9.55VVDD115 pKa = 5.51RR116 pKa = 11.84ISPVILSAPPEE127 pKa = 4.16SDD129 pKa = 4.72PRR131 pKa = 11.84LEE133 pKa = 4.47WIASYY138 pKa = 9.73WLNLLVIYY146 pKa = 9.62SPPLARR152 pKa = 11.84SIRR155 pKa = 11.84AWALAGEE162 pKa = 4.35NPLYY166 pKa = 10.15TLQEE170 pKa = 4.22VQEE173 pKa = 4.78KK174 pKa = 10.11YY175 pKa = 10.44PPAQGTVVTPIAPLHH190 pKa = 5.3QLIHH194 pKa = 6.55TYY196 pKa = 10.54FAMTLGFKK204 pKa = 9.99NVNSDD209 pKa = 4.68YY210 pKa = 11.09ISDD213 pKa = 3.88WAQKK217 pKa = 9.85RR218 pKa = 11.84ANSMLAGLGLDD229 pKa = 3.7KK230 pKa = 11.13ASASAAIAGNFPTAAVTSTFFQSHH254 pKa = 5.61PTLLRR259 pKa = 11.84FIIDD263 pKa = 4.03FVTANLTEE271 pKa = 4.68CEE273 pKa = 4.19DD274 pKa = 3.78PGIQKK279 pKa = 10.22LCSIVDD285 pKa = 4.11MILNKK290 pKa = 10.62AGMATIEE297 pKa = 4.17SAYY300 pKa = 10.71NFAKK304 pKa = 10.49AASTAASFLPRR315 pKa = 11.84VAEE318 pKa = 3.79QSVALIEE325 pKa = 3.89IWEE328 pKa = 4.13AMMKK332 pKa = 10.72DD333 pKa = 3.55EE334 pKa = 5.81GEE336 pKa = 4.48VTQTAAGNVHH346 pKa = 6.5GLGAMFPYY354 pKa = 10.43ARR356 pKa = 11.84ALGLRR361 pKa = 11.84GVEE364 pKa = 3.9LLNVGKK370 pKa = 10.42YY371 pKa = 10.66ADD373 pKa = 4.06LAYY376 pKa = 10.46CAWFQRR382 pKa = 11.84ADD384 pKa = 3.6EE385 pKa = 4.42NPTYY389 pKa = 11.14RR390 pKa = 11.84NMAHH394 pKa = 6.38SQATDD399 pKa = 3.16QKK401 pKa = 9.72TLAAHH406 pKa = 6.08TKK408 pKa = 8.38VPLSIGYY415 pKa = 7.67TSLTEE420 pKa = 4.15TVKK423 pKa = 10.87KK424 pKa = 10.53FLDD427 pKa = 3.36EE428 pKa = 4.48RR429 pKa = 11.84GISEE433 pKa = 3.92AQYY436 pKa = 11.1TEE438 pKa = 4.07GCKK441 pKa = 9.52QAAKK445 pKa = 10.57NMEE448 pKa = 4.63LIKK451 pKa = 10.7PEE453 pKa = 3.85EE454 pKa = 3.87LRR456 pKa = 11.84SNFEE460 pKa = 3.99LLTRR464 pKa = 11.84RR465 pKa = 3.98
MM1 pKa = 7.92DD2 pKa = 4.8TFQIDD7 pKa = 3.76PASDD11 pKa = 3.71TPCPHH16 pKa = 6.92RR17 pKa = 11.84AIVFQPAGGRR27 pKa = 11.84SVRR30 pKa = 11.84IYY32 pKa = 10.97ARR34 pKa = 11.84PPDD37 pKa = 3.66QGAAPSYY44 pKa = 10.14PIYY47 pKa = 10.55LCPNADD53 pKa = 3.82FLTLSVDD60 pKa = 3.03ALTIGGYY67 pKa = 9.99GDD69 pKa = 3.56HH70 pKa = 6.21VQYY73 pKa = 10.81DD74 pKa = 3.51IRR76 pKa = 11.84NQIEE80 pKa = 4.0PQDD83 pKa = 3.77WVADD87 pKa = 3.75SATTFTVAPSRR98 pKa = 11.84LPVWIGIDD106 pKa = 3.29HH107 pKa = 7.17AAAKK111 pKa = 10.11YY112 pKa = 9.55VVDD115 pKa = 5.51RR116 pKa = 11.84ISPVILSAPPEE127 pKa = 4.16SDD129 pKa = 4.72PRR131 pKa = 11.84LEE133 pKa = 4.47WIASYY138 pKa = 9.73WLNLLVIYY146 pKa = 9.62SPPLARR152 pKa = 11.84SIRR155 pKa = 11.84AWALAGEE162 pKa = 4.35NPLYY166 pKa = 10.15TLQEE170 pKa = 4.22VQEE173 pKa = 4.78KK174 pKa = 10.11YY175 pKa = 10.44PPAQGTVVTPIAPLHH190 pKa = 5.3QLIHH194 pKa = 6.55TYY196 pKa = 10.54FAMTLGFKK204 pKa = 9.99NVNSDD209 pKa = 4.68YY210 pKa = 11.09ISDD213 pKa = 3.88WAQKK217 pKa = 9.85RR218 pKa = 11.84ANSMLAGLGLDD229 pKa = 3.7KK230 pKa = 11.13ASASAAIAGNFPTAAVTSTFFQSHH254 pKa = 5.61PTLLRR259 pKa = 11.84FIIDD263 pKa = 4.03FVTANLTEE271 pKa = 4.68CEE273 pKa = 4.19DD274 pKa = 3.78PGIQKK279 pKa = 10.22LCSIVDD285 pKa = 4.11MILNKK290 pKa = 10.62AGMATIEE297 pKa = 4.17SAYY300 pKa = 10.71NFAKK304 pKa = 10.49AASTAASFLPRR315 pKa = 11.84VAEE318 pKa = 3.79QSVALIEE325 pKa = 3.89IWEE328 pKa = 4.13AMMKK332 pKa = 10.72DD333 pKa = 3.55EE334 pKa = 5.81GEE336 pKa = 4.48VTQTAAGNVHH346 pKa = 6.5GLGAMFPYY354 pKa = 10.43ARR356 pKa = 11.84ALGLRR361 pKa = 11.84GVEE364 pKa = 3.9LLNVGKK370 pKa = 10.42YY371 pKa = 10.66ADD373 pKa = 4.06LAYY376 pKa = 10.46CAWFQRR382 pKa = 11.84ADD384 pKa = 3.6EE385 pKa = 4.42NPTYY389 pKa = 11.14RR390 pKa = 11.84NMAHH394 pKa = 6.38SQATDD399 pKa = 3.16QKK401 pKa = 9.72TLAAHH406 pKa = 6.08TKK408 pKa = 8.38VPLSIGYY415 pKa = 7.67TSLTEE420 pKa = 4.15TVKK423 pKa = 10.87KK424 pKa = 10.53FLDD427 pKa = 3.36EE428 pKa = 4.48RR429 pKa = 11.84GISEE433 pKa = 3.92AQYY436 pKa = 11.1TEE438 pKa = 4.07GCKK441 pKa = 9.52QAAKK445 pKa = 10.57NMEE448 pKa = 4.63LIKK451 pKa = 10.7PEE453 pKa = 3.85EE454 pKa = 3.87LRR456 pKa = 11.84SNFEE460 pKa = 3.99LLTRR464 pKa = 11.84RR465 pKa = 3.98
Molecular weight: 51.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KNA8|A0A1L3KNA8_9MONO Uncharacterized protein OS=Hubei orthoptera virus 5 OX=1923013 PE=4 SV=1
MM1 pKa = 7.5AGKK4 pKa = 9.67FVFGLTVSILVSFISGRR21 pKa = 11.84FVSKK25 pKa = 10.98HH26 pKa = 4.4EE27 pKa = 3.9VAIIADD33 pKa = 3.95EE34 pKa = 4.32FTTVTLGVKK43 pKa = 9.7ILMPSDD49 pKa = 3.63LVKK52 pKa = 10.81DD53 pKa = 3.86ILTSLPDD60 pKa = 3.55NAPPATHH67 pKa = 6.93EE68 pKa = 4.25GRR70 pKa = 11.84AKK72 pKa = 10.27LVKK75 pKa = 10.23LLSEE79 pKa = 4.58LYY81 pKa = 10.3DD82 pKa = 3.62RR83 pKa = 11.84SLRR86 pKa = 11.84RR87 pKa = 11.84EE88 pKa = 3.99MINVNKK94 pKa = 10.36AVTHH98 pKa = 6.42DD99 pKa = 4.11LSEE102 pKa = 4.39DD103 pKa = 3.59SYY105 pKa = 11.29MSNLTLHH112 pKa = 7.08ASCTYY117 pKa = 10.32FPDD120 pKa = 4.05SNSSGDD126 pKa = 3.61CSNISMGSDD135 pKa = 3.07VRR137 pKa = 11.84PPPKK141 pKa = 9.22TDD143 pKa = 3.05HH144 pKa = 6.96LEE146 pKa = 4.15LYY148 pKa = 10.26LPRR151 pKa = 11.84EE152 pKa = 4.17LRR154 pKa = 11.84STRR157 pKa = 11.84RR158 pKa = 11.84KK159 pKa = 9.47RR160 pKa = 11.84SSDD163 pKa = 3.08VVSYY167 pKa = 10.89MNTLLNLRR175 pKa = 11.84SAPDD179 pKa = 3.55EE180 pKa = 4.31YY181 pKa = 11.38SCSTLIPCIYY191 pKa = 9.9HH192 pKa = 5.45LTIGQFTYY200 pKa = 10.65LYY202 pKa = 9.98IVPQGDD208 pKa = 3.59ASCPSICEE216 pKa = 4.17DD217 pKa = 3.46GVSIGVLVASTTDD230 pKa = 3.4PSYY233 pKa = 11.13SWRR236 pKa = 11.84FGNQILACGTRR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84NSLLCSIFPRR259 pKa = 11.84QHH261 pKa = 5.48TCTCEE266 pKa = 3.8VLIQANNNLITPRR279 pKa = 11.84TIRR282 pKa = 11.84RR283 pKa = 11.84GKK285 pKa = 10.27RR286 pKa = 11.84GVTSGLRR293 pKa = 11.84RR294 pKa = 11.84AFNWLSKK301 pKa = 8.85VLKK304 pKa = 9.49WGPKK308 pKa = 9.68KK309 pKa = 10.08ILHH312 pKa = 5.9SFKK315 pKa = 9.85KK316 pKa = 10.18HH317 pKa = 4.36PWRR320 pKa = 11.84TVGGTVGFASGVYY333 pKa = 9.27TIYY336 pKa = 11.0EE337 pKa = 4.12LVEE340 pKa = 4.49LKK342 pKa = 10.66LSPPVPSHH350 pKa = 5.95EE351 pKa = 4.53EE352 pKa = 3.43IVSIVEE358 pKa = 4.22KK359 pKa = 10.32MRR361 pKa = 11.84QTDD364 pKa = 3.4QALVNVSDD372 pKa = 3.9SLYY375 pKa = 9.24VGIGAVHH382 pKa = 7.71DD383 pKa = 4.36DD384 pKa = 4.24LLNFRR389 pKa = 11.84QDD391 pKa = 3.36MKK393 pKa = 11.5DD394 pKa = 3.06SFHH397 pKa = 7.29QLGQWSDD404 pKa = 3.62QVTVIHH410 pKa = 6.52TLLSLCVSRR419 pKa = 11.84GQSGLSGLLRR429 pKa = 11.84GEE431 pKa = 4.55LPQTVISQEE440 pKa = 4.04SLQLFISTLLGKK452 pKa = 9.65EE453 pKa = 3.99LSSAVAMMPLPIRR466 pKa = 11.84LVNADD471 pKa = 5.25FISNMVWVSVRR482 pKa = 11.84VPKK485 pKa = 10.69KK486 pKa = 9.45GTTLLGRR493 pKa = 11.84VYY495 pKa = 10.64RR496 pKa = 11.84LDD498 pKa = 3.68GEE500 pKa = 4.58TRR502 pKa = 11.84TVNTPNDD509 pKa = 2.79ICYY512 pKa = 7.28TTRR515 pKa = 11.84GDD517 pKa = 3.42QYY519 pKa = 11.21YY520 pKa = 9.47FAPTSPDD527 pKa = 2.89IKK529 pKa = 9.12PTPSHH534 pKa = 7.21RR535 pKa = 11.84ITLEE539 pKa = 3.63SCTKK543 pKa = 10.29SGGILICPYY552 pKa = 11.07GSLTDD557 pKa = 3.51VTKK560 pKa = 8.73TTEE563 pKa = 3.59HH564 pKa = 7.37DD565 pKa = 3.38ICFSRR570 pKa = 11.84YY571 pKa = 9.3SSPKK575 pKa = 8.88RR576 pKa = 11.84SRR578 pKa = 11.84RR579 pKa = 11.84DD580 pKa = 3.26VNKK583 pKa = 10.62DD584 pKa = 2.64IITEE588 pKa = 4.08VLRR591 pKa = 11.84KK592 pKa = 8.35TSTFTEE598 pKa = 4.22IPLPSDD604 pKa = 3.5TPPSTFHH611 pKa = 6.62QFTEE615 pKa = 4.68RR616 pKa = 11.84YY617 pKa = 8.32KK618 pKa = 10.75VAISSLQEE626 pKa = 3.85SNKK629 pKa = 9.97RR630 pKa = 11.84LEE632 pKa = 4.64DD633 pKa = 3.05AWQEE637 pKa = 3.95LLEE640 pKa = 4.28KK641 pKa = 10.6HH642 pKa = 6.44SSLNLWSRR650 pKa = 11.84LHH652 pKa = 6.28LAWTSVFSFLTLLLVVKK669 pKa = 9.66VGWDD673 pKa = 2.78ILTTRR678 pKa = 11.84RR679 pKa = 11.84FRR681 pKa = 11.84QGLKK685 pKa = 8.82VTPDD689 pKa = 2.76HH690 pKa = 6.87HH691 pKa = 6.37EE692 pKa = 4.03AVSLRR697 pKa = 11.84EE698 pKa = 4.07RR699 pKa = 11.84GCDD702 pKa = 3.46TII704 pKa = 5.3
MM1 pKa = 7.5AGKK4 pKa = 9.67FVFGLTVSILVSFISGRR21 pKa = 11.84FVSKK25 pKa = 10.98HH26 pKa = 4.4EE27 pKa = 3.9VAIIADD33 pKa = 3.95EE34 pKa = 4.32FTTVTLGVKK43 pKa = 9.7ILMPSDD49 pKa = 3.63LVKK52 pKa = 10.81DD53 pKa = 3.86ILTSLPDD60 pKa = 3.55NAPPATHH67 pKa = 6.93EE68 pKa = 4.25GRR70 pKa = 11.84AKK72 pKa = 10.27LVKK75 pKa = 10.23LLSEE79 pKa = 4.58LYY81 pKa = 10.3DD82 pKa = 3.62RR83 pKa = 11.84SLRR86 pKa = 11.84RR87 pKa = 11.84EE88 pKa = 3.99MINVNKK94 pKa = 10.36AVTHH98 pKa = 6.42DD99 pKa = 4.11LSEE102 pKa = 4.39DD103 pKa = 3.59SYY105 pKa = 11.29MSNLTLHH112 pKa = 7.08ASCTYY117 pKa = 10.32FPDD120 pKa = 4.05SNSSGDD126 pKa = 3.61CSNISMGSDD135 pKa = 3.07VRR137 pKa = 11.84PPPKK141 pKa = 9.22TDD143 pKa = 3.05HH144 pKa = 6.96LEE146 pKa = 4.15LYY148 pKa = 10.26LPRR151 pKa = 11.84EE152 pKa = 4.17LRR154 pKa = 11.84STRR157 pKa = 11.84RR158 pKa = 11.84KK159 pKa = 9.47RR160 pKa = 11.84SSDD163 pKa = 3.08VVSYY167 pKa = 10.89MNTLLNLRR175 pKa = 11.84SAPDD179 pKa = 3.55EE180 pKa = 4.31YY181 pKa = 11.38SCSTLIPCIYY191 pKa = 9.9HH192 pKa = 5.45LTIGQFTYY200 pKa = 10.65LYY202 pKa = 9.98IVPQGDD208 pKa = 3.59ASCPSICEE216 pKa = 4.17DD217 pKa = 3.46GVSIGVLVASTTDD230 pKa = 3.4PSYY233 pKa = 11.13SWRR236 pKa = 11.84FGNQILACGTRR247 pKa = 11.84RR248 pKa = 11.84RR249 pKa = 11.84NSLLCSIFPRR259 pKa = 11.84QHH261 pKa = 5.48TCTCEE266 pKa = 3.8VLIQANNNLITPRR279 pKa = 11.84TIRR282 pKa = 11.84RR283 pKa = 11.84GKK285 pKa = 10.27RR286 pKa = 11.84GVTSGLRR293 pKa = 11.84RR294 pKa = 11.84AFNWLSKK301 pKa = 8.85VLKK304 pKa = 9.49WGPKK308 pKa = 9.68KK309 pKa = 10.08ILHH312 pKa = 5.9SFKK315 pKa = 9.85KK316 pKa = 10.18HH317 pKa = 4.36PWRR320 pKa = 11.84TVGGTVGFASGVYY333 pKa = 9.27TIYY336 pKa = 11.0EE337 pKa = 4.12LVEE340 pKa = 4.49LKK342 pKa = 10.66LSPPVPSHH350 pKa = 5.95EE351 pKa = 4.53EE352 pKa = 3.43IVSIVEE358 pKa = 4.22KK359 pKa = 10.32MRR361 pKa = 11.84QTDD364 pKa = 3.4QALVNVSDD372 pKa = 3.9SLYY375 pKa = 9.24VGIGAVHH382 pKa = 7.71DD383 pKa = 4.36DD384 pKa = 4.24LLNFRR389 pKa = 11.84QDD391 pKa = 3.36MKK393 pKa = 11.5DD394 pKa = 3.06SFHH397 pKa = 7.29QLGQWSDD404 pKa = 3.62QVTVIHH410 pKa = 6.52TLLSLCVSRR419 pKa = 11.84GQSGLSGLLRR429 pKa = 11.84GEE431 pKa = 4.55LPQTVISQEE440 pKa = 4.04SLQLFISTLLGKK452 pKa = 9.65EE453 pKa = 3.99LSSAVAMMPLPIRR466 pKa = 11.84LVNADD471 pKa = 5.25FISNMVWVSVRR482 pKa = 11.84VPKK485 pKa = 10.69KK486 pKa = 9.45GTTLLGRR493 pKa = 11.84VYY495 pKa = 10.64RR496 pKa = 11.84LDD498 pKa = 3.68GEE500 pKa = 4.58TRR502 pKa = 11.84TVNTPNDD509 pKa = 2.79ICYY512 pKa = 7.28TTRR515 pKa = 11.84GDD517 pKa = 3.42QYY519 pKa = 11.21YY520 pKa = 9.47FAPTSPDD527 pKa = 2.89IKK529 pKa = 9.12PTPSHH534 pKa = 7.21RR535 pKa = 11.84ITLEE539 pKa = 3.63SCTKK543 pKa = 10.29SGGILICPYY552 pKa = 11.07GSLTDD557 pKa = 3.51VTKK560 pKa = 8.73TTEE563 pKa = 3.59HH564 pKa = 7.37DD565 pKa = 3.38ICFSRR570 pKa = 11.84YY571 pKa = 9.3SSPKK575 pKa = 8.88RR576 pKa = 11.84SRR578 pKa = 11.84RR579 pKa = 11.84DD580 pKa = 3.26VNKK583 pKa = 10.62DD584 pKa = 2.64IITEE588 pKa = 4.08VLRR591 pKa = 11.84KK592 pKa = 8.35TSTFTEE598 pKa = 4.22IPLPSDD604 pKa = 3.5TPPSTFHH611 pKa = 6.62QFTEE615 pKa = 4.68RR616 pKa = 11.84YY617 pKa = 8.32KK618 pKa = 10.75VAISSLQEE626 pKa = 3.85SNKK629 pKa = 9.97RR630 pKa = 11.84LEE632 pKa = 4.64DD633 pKa = 3.05AWQEE637 pKa = 3.95LLEE640 pKa = 4.28KK641 pKa = 10.6HH642 pKa = 6.44SSLNLWSRR650 pKa = 11.84LHH652 pKa = 6.28LAWTSVFSFLTLLLVVKK669 pKa = 9.66VGWDD673 pKa = 2.78ILTTRR678 pKa = 11.84RR679 pKa = 11.84FRR681 pKa = 11.84QGLKK685 pKa = 8.82VTPDD689 pKa = 2.76HH690 pKa = 6.87HH691 pKa = 6.37EE692 pKa = 4.03AVSLRR697 pKa = 11.84EE698 pKa = 4.07RR699 pKa = 11.84GCDD702 pKa = 3.46TII704 pKa = 5.3
Molecular weight: 78.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3383 |
288 |
1926 |
845.8 |
93.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.124 ± 1.544 | 2.01 ± 0.279 |
4.877 ± 0.29 | 4.73 ± 0.194 |
3.636 ± 0.183 | 6.592 ± 0.523 |
2.424 ± 0.207 | 5.616 ± 0.52 |
4.523 ± 0.309 | 11.558 ± 0.72 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.069 ± 0.181 | 3.163 ± 0.129 |
5.823 ± 0.487 | 3.458 ± 0.328 |
5.616 ± 0.481 | 8.75 ± 1.111 |
6.71 ± 0.554 | 6.562 ± 0.404 |
1.567 ± 0.202 | 3.192 ± 0.271 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
