
Porcine associated porprismacovirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
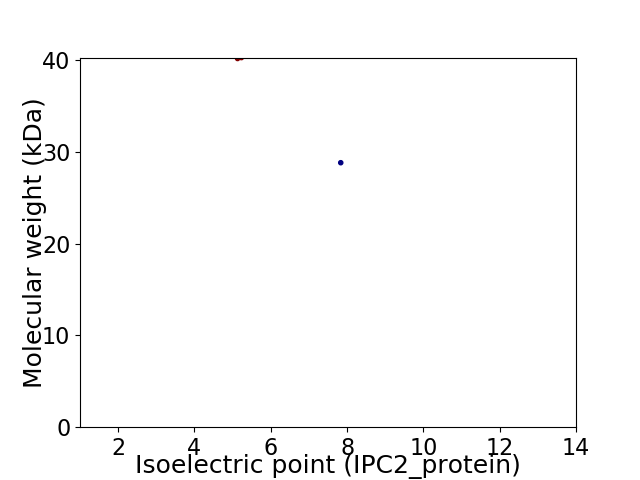
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A076VEN2|A0A076VEN2_9VIRU Cap protein OS=Porcine associated porprismacovirus 5 OX=2170121 PE=4 SV=1
MM1 pKa = 6.78ATNYY5 pKa = 7.69VTAKK9 pKa = 9.39YY10 pKa = 10.33QEE12 pKa = 4.47IYY14 pKa = 11.05DD15 pKa = 4.59LGTQAGKK22 pKa = 7.12TTILGIHH29 pKa = 6.11TPDD32 pKa = 3.53GGNVYY37 pKa = 11.15NMLSGFFDD45 pKa = 3.12QFRR48 pKa = 11.84KK49 pKa = 10.1YY50 pKa = 9.98RR51 pKa = 11.84YY52 pKa = 8.46KK53 pKa = 9.8GCSVSMVPAAQLPADD68 pKa = 4.06PLQVSFEE75 pKa = 4.28AGALTCDD82 pKa = 4.45PRR84 pKa = 11.84DD85 pKa = 3.67LLNPILFHH93 pKa = 6.36GCHH96 pKa = 5.99GEE98 pKa = 3.96EE99 pKa = 4.42LNEE102 pKa = 4.19ILDD105 pKa = 3.73SVYY108 pKa = 11.28YY109 pKa = 10.58KK110 pKa = 10.04GTSPSSVRR118 pKa = 11.84EE119 pKa = 4.05LQVNTQVSEE128 pKa = 3.91ASYY131 pKa = 10.76YY132 pKa = 10.88AALSDD137 pKa = 3.46PSFRR141 pKa = 11.84KK142 pKa = 9.93FGIQQGAKK150 pKa = 8.93VSLRR154 pKa = 11.84PMVHH158 pKa = 6.81PLAVTQPIEE167 pKa = 4.26HH168 pKa = 6.98SDD170 pKa = 3.01SKK172 pKa = 11.41YY173 pKa = 9.82YY174 pKa = 10.81YY175 pKa = 9.32SYY177 pKa = 10.8PSGGTTVTGSVDD189 pKa = 3.36GVEE192 pKa = 4.38SGDD195 pKa = 3.93LGMVQNGTNFTRR207 pKa = 11.84FGLQDD212 pKa = 3.13ACDD215 pKa = 3.63AVEE218 pKa = 4.1LHH220 pKa = 6.34VSGAGIPSGSVYY232 pKa = 10.59VSPTKK237 pKa = 10.41TIFSNGLKK245 pKa = 9.64PLSWLPTWHH254 pKa = 6.3FVPYY258 pKa = 10.24AGEE261 pKa = 3.99NMLTVPYY268 pKa = 9.78QNRR271 pKa = 11.84VSDD274 pKa = 3.59MSIEE278 pKa = 4.29TTRR281 pKa = 11.84LPLLYY286 pKa = 9.15MGLFILPPSYY296 pKa = 9.24TQEE299 pKa = 3.69MYY301 pKa = 10.28FRR303 pKa = 11.84LIITHH308 pKa = 5.6YY309 pKa = 11.04FEE311 pKa = 6.03FKK313 pKa = 10.81DD314 pKa = 4.68FSTCLTRR321 pKa = 11.84HH322 pKa = 5.99SVDD325 pKa = 3.2QLNPDD330 pKa = 3.21RR331 pKa = 11.84KK332 pKa = 10.39VYY334 pKa = 10.5TEE336 pKa = 4.8TIPEE340 pKa = 4.16PTSGSSASSLTVVDD354 pKa = 5.48GSAVQTSEE362 pKa = 3.93GVYY365 pKa = 10.53
MM1 pKa = 6.78ATNYY5 pKa = 7.69VTAKK9 pKa = 9.39YY10 pKa = 10.33QEE12 pKa = 4.47IYY14 pKa = 11.05DD15 pKa = 4.59LGTQAGKK22 pKa = 7.12TTILGIHH29 pKa = 6.11TPDD32 pKa = 3.53GGNVYY37 pKa = 11.15NMLSGFFDD45 pKa = 3.12QFRR48 pKa = 11.84KK49 pKa = 10.1YY50 pKa = 9.98RR51 pKa = 11.84YY52 pKa = 8.46KK53 pKa = 9.8GCSVSMVPAAQLPADD68 pKa = 4.06PLQVSFEE75 pKa = 4.28AGALTCDD82 pKa = 4.45PRR84 pKa = 11.84DD85 pKa = 3.67LLNPILFHH93 pKa = 6.36GCHH96 pKa = 5.99GEE98 pKa = 3.96EE99 pKa = 4.42LNEE102 pKa = 4.19ILDD105 pKa = 3.73SVYY108 pKa = 11.28YY109 pKa = 10.58KK110 pKa = 10.04GTSPSSVRR118 pKa = 11.84EE119 pKa = 4.05LQVNTQVSEE128 pKa = 3.91ASYY131 pKa = 10.76YY132 pKa = 10.88AALSDD137 pKa = 3.46PSFRR141 pKa = 11.84KK142 pKa = 9.93FGIQQGAKK150 pKa = 8.93VSLRR154 pKa = 11.84PMVHH158 pKa = 6.81PLAVTQPIEE167 pKa = 4.26HH168 pKa = 6.98SDD170 pKa = 3.01SKK172 pKa = 11.41YY173 pKa = 9.82YY174 pKa = 10.81YY175 pKa = 9.32SYY177 pKa = 10.8PSGGTTVTGSVDD189 pKa = 3.36GVEE192 pKa = 4.38SGDD195 pKa = 3.93LGMVQNGTNFTRR207 pKa = 11.84FGLQDD212 pKa = 3.13ACDD215 pKa = 3.63AVEE218 pKa = 4.1LHH220 pKa = 6.34VSGAGIPSGSVYY232 pKa = 10.59VSPTKK237 pKa = 10.41TIFSNGLKK245 pKa = 9.64PLSWLPTWHH254 pKa = 6.3FVPYY258 pKa = 10.24AGEE261 pKa = 3.99NMLTVPYY268 pKa = 9.78QNRR271 pKa = 11.84VSDD274 pKa = 3.59MSIEE278 pKa = 4.29TTRR281 pKa = 11.84LPLLYY286 pKa = 9.15MGLFILPPSYY296 pKa = 9.24TQEE299 pKa = 3.69MYY301 pKa = 10.28FRR303 pKa = 11.84LIITHH308 pKa = 5.6YY309 pKa = 11.04FEE311 pKa = 6.03FKK313 pKa = 10.81DD314 pKa = 4.68FSTCLTRR321 pKa = 11.84HH322 pKa = 5.99SVDD325 pKa = 3.2QLNPDD330 pKa = 3.21RR331 pKa = 11.84KK332 pKa = 10.39VYY334 pKa = 10.5TEE336 pKa = 4.8TIPEE340 pKa = 4.16PTSGSSASSLTVVDD354 pKa = 5.48GSAVQTSEE362 pKa = 3.93GVYY365 pKa = 10.53
Molecular weight: 40.14 kDa
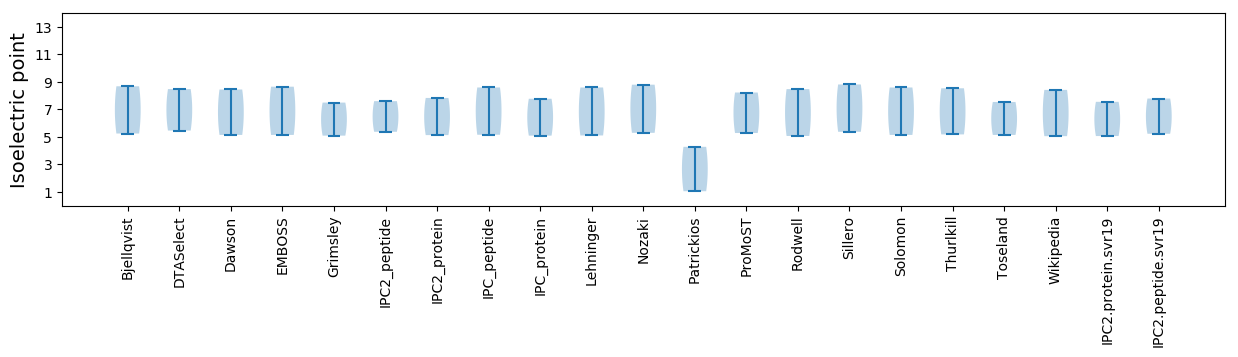
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076VEN2|A0A076VEN2_9VIRU Cap protein OS=Porcine associated porprismacovirus 5 OX=2170121 PE=4 SV=1
MM1 pKa = 6.82QAYY4 pKa = 9.2MMTIPRR10 pKa = 11.84TVSKK14 pKa = 10.35RR15 pKa = 11.84ALRR18 pKa = 11.84IMIDD22 pKa = 3.53VNDD25 pKa = 3.98CKK27 pKa = 10.84KK28 pKa = 10.0WIIGKK33 pKa = 9.24EE34 pKa = 3.97EE35 pKa = 3.89GKK37 pKa = 10.27NGYY40 pKa = 8.24KK41 pKa = 9.71HH42 pKa = 4.18WQIRR46 pKa = 11.84IEE48 pKa = 4.05TSNDD52 pKa = 3.05SFFEE56 pKa = 3.95WMQDD60 pKa = 3.65HH61 pKa = 7.09IPTAHH66 pKa = 7.41IEE68 pKa = 4.07RR69 pKa = 11.84TEE71 pKa = 4.27CGVDD75 pKa = 2.94ACRR78 pKa = 11.84YY79 pKa = 5.85EE80 pKa = 4.53AKK82 pKa = 10.15EE83 pKa = 4.03GQYY86 pKa = 11.57VMYY89 pKa = 10.37SDD91 pKa = 5.32RR92 pKa = 11.84PQNLMQRR99 pKa = 11.84YY100 pKa = 9.44GEE102 pKa = 3.85FRR104 pKa = 11.84PNQKK108 pKa = 9.86HH109 pKa = 6.04ALQALQQSNDD119 pKa = 3.33RR120 pKa = 11.84EE121 pKa = 4.4VVVWYY126 pKa = 10.26DD127 pKa = 3.15EE128 pKa = 4.19TGNVGKK134 pKa = 10.24SWFTGALWEE143 pKa = 4.29RR144 pKa = 11.84KK145 pKa = 8.78LAYY148 pKa = 7.69VTPPTIDD155 pKa = 3.16TVKK158 pKa = 11.11GLIQWIASCYY168 pKa = 9.96IEE170 pKa = 5.69DD171 pKa = 3.38GWRR174 pKa = 11.84PYY176 pKa = 10.39IIIDD180 pKa = 4.57IPRR183 pKa = 11.84SWKK186 pKa = 9.67WSEE189 pKa = 3.8QLYY192 pKa = 10.74SAIEE196 pKa = 4.22SIKK199 pKa = 11.0DD200 pKa = 3.09GLIYY204 pKa = 9.09DD205 pKa = 4.14TRR207 pKa = 11.84YY208 pKa = 9.36HH209 pKa = 7.23SSMMNIRR216 pKa = 11.84GVKK219 pKa = 9.02VLVMTNTMPKK229 pKa = 9.86LDD231 pKa = 4.6KK232 pKa = 10.62LSKK235 pKa = 10.59DD236 pKa = 2.97RR237 pKa = 11.84WCIRR241 pKa = 11.84TFF243 pKa = 3.22
MM1 pKa = 6.82QAYY4 pKa = 9.2MMTIPRR10 pKa = 11.84TVSKK14 pKa = 10.35RR15 pKa = 11.84ALRR18 pKa = 11.84IMIDD22 pKa = 3.53VNDD25 pKa = 3.98CKK27 pKa = 10.84KK28 pKa = 10.0WIIGKK33 pKa = 9.24EE34 pKa = 3.97EE35 pKa = 3.89GKK37 pKa = 10.27NGYY40 pKa = 8.24KK41 pKa = 9.71HH42 pKa = 4.18WQIRR46 pKa = 11.84IEE48 pKa = 4.05TSNDD52 pKa = 3.05SFFEE56 pKa = 3.95WMQDD60 pKa = 3.65HH61 pKa = 7.09IPTAHH66 pKa = 7.41IEE68 pKa = 4.07RR69 pKa = 11.84TEE71 pKa = 4.27CGVDD75 pKa = 2.94ACRR78 pKa = 11.84YY79 pKa = 5.85EE80 pKa = 4.53AKK82 pKa = 10.15EE83 pKa = 4.03GQYY86 pKa = 11.57VMYY89 pKa = 10.37SDD91 pKa = 5.32RR92 pKa = 11.84PQNLMQRR99 pKa = 11.84YY100 pKa = 9.44GEE102 pKa = 3.85FRR104 pKa = 11.84PNQKK108 pKa = 9.86HH109 pKa = 6.04ALQALQQSNDD119 pKa = 3.33RR120 pKa = 11.84EE121 pKa = 4.4VVVWYY126 pKa = 10.26DD127 pKa = 3.15EE128 pKa = 4.19TGNVGKK134 pKa = 10.24SWFTGALWEE143 pKa = 4.29RR144 pKa = 11.84KK145 pKa = 8.78LAYY148 pKa = 7.69VTPPTIDD155 pKa = 3.16TVKK158 pKa = 11.11GLIQWIASCYY168 pKa = 9.96IEE170 pKa = 5.69DD171 pKa = 3.38GWRR174 pKa = 11.84PYY176 pKa = 10.39IIIDD180 pKa = 4.57IPRR183 pKa = 11.84SWKK186 pKa = 9.67WSEE189 pKa = 3.8QLYY192 pKa = 10.74SAIEE196 pKa = 4.22SIKK199 pKa = 11.0DD200 pKa = 3.09GLIYY204 pKa = 9.09DD205 pKa = 4.14TRR207 pKa = 11.84YY208 pKa = 9.36HH209 pKa = 7.23SSMMNIRR216 pKa = 11.84GVKK219 pKa = 9.02VLVMTNTMPKK229 pKa = 9.86LDD231 pKa = 4.6KK232 pKa = 10.62LSKK235 pKa = 10.59DD236 pKa = 2.97RR237 pKa = 11.84WCIRR241 pKa = 11.84TFF243 pKa = 3.22
Molecular weight: 28.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
608 |
243 |
365 |
304.0 |
34.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.934 ± 0.236 | 1.645 ± 0.24 |
5.428 ± 0.432 | 5.263 ± 0.528 |
3.454 ± 0.81 | 7.072 ± 1.0 |
2.303 ± 0.142 | 5.921 ± 1.818 |
4.77 ± 1.292 | 7.072 ± 1.238 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.289 ± 0.718 | 3.454 ± 0.145 |
5.428 ± 1.0 | 4.605 ± 0.193 |
4.77 ± 1.292 | 8.388 ± 1.524 |
7.237 ± 0.856 | 6.908 ± 0.904 |
2.138 ± 1.386 | 5.921 ± 0.332 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
