
Jonesia denitrificans (strain ATCC 14870 / DSM 20603 / BCRC 15368 / CIP 55.134 / JCM 11481 / NBRC 15587 / NCTC 10816 / Prevot 55134) (Listeria denitrificans)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Jonesiaceae; Jonesia; Jonesia denitrificans
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
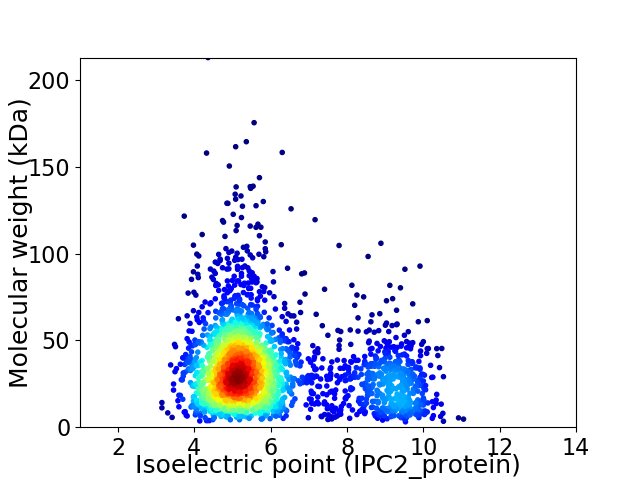
Virtual 2D-PAGE plot for 2495 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7QZC1|C7QZC1_JONDD ATP synthase subunit alpha OS=Jonesia denitrificans (strain ATCC 14870 / DSM 20603 / BCRC 15368 / CIP 55.134 / JCM 11481 / NBRC 15587 / NCTC 10816 / Prevot 55134) OX=471856 GN=atpA PE=3 SV=1
MM1 pKa = 6.95NHH3 pKa = 5.75HH4 pKa = 6.59TATHH8 pKa = 5.32TQATRR13 pKa = 11.84RR14 pKa = 11.84PLTTTLTALTLTAALAGCVPGSSNEE39 pKa = 3.98DD40 pKa = 3.24NAQGEE45 pKa = 4.74TPSTVTVVVHH55 pKa = 7.28DD56 pKa = 4.16SFQMSEE62 pKa = 3.93EE63 pKa = 3.93QQADD67 pKa = 3.73FEE69 pKa = 4.63KK70 pKa = 10.55TSGYY74 pKa = 11.06DD75 pKa = 3.2LTIVTNGDD83 pKa = 2.76GGALVNSLILTKK95 pKa = 10.44DD96 pKa = 3.19APIADD101 pKa = 3.46AVFGVDD107 pKa = 2.9NTFASRR113 pKa = 11.84GISEE117 pKa = 4.79EE118 pKa = 3.79VFAPYY123 pKa = 9.74TSEE126 pKa = 4.13VASQTSRR133 pKa = 11.84DD134 pKa = 3.49LALPGDD140 pKa = 4.23DD141 pKa = 4.09TLTAIDD147 pKa = 4.99FGDD150 pKa = 3.29VCLNVDD156 pKa = 3.59PTWFEE161 pKa = 3.8NHH163 pKa = 6.31SLDD166 pKa = 5.04VPATLDD172 pKa = 3.75DD173 pKa = 4.67LADD176 pKa = 3.7PAYY179 pKa = 10.88RR180 pKa = 11.84NLTVVTNPATSSPGLSFLFATIGKK204 pKa = 9.13YY205 pKa = 10.71GEE207 pKa = 5.08DD208 pKa = 3.88GYY210 pKa = 11.56LDD212 pKa = 3.67YY213 pKa = 8.34WTQLKK218 pKa = 11.03DD219 pKa = 3.35NGLKK223 pKa = 9.97IVDD226 pKa = 3.86GWEE229 pKa = 3.39DD230 pKa = 3.24AYY232 pKa = 11.66YY233 pKa = 11.24VDD235 pKa = 5.62FTATGDD241 pKa = 3.5GDD243 pKa = 3.91RR244 pKa = 11.84PIALSYY250 pKa = 10.85ASSPAFTVTDD260 pKa = 4.39DD261 pKa = 3.96GSATTTSALLDD272 pKa = 3.36TCFRR276 pKa = 11.84QVEE279 pKa = 4.46YY280 pKa = 11.15AGVLHH285 pKa = 6.87GAANPEE291 pKa = 3.98GAQAFIDD298 pKa = 3.87YY299 pKa = 10.22LLSEE303 pKa = 5.57DD304 pKa = 4.22YY305 pKa = 9.65QTTIPDD311 pKa = 3.58AMYY314 pKa = 8.45MYY316 pKa = 10.21PVADD320 pKa = 3.61TVEE323 pKa = 5.4LPDD326 pKa = 3.69AWAQFAPVPTTPISLDD342 pKa = 3.33PDD344 pKa = 4.21TIARR348 pKa = 11.84MRR350 pKa = 11.84DD351 pKa = 2.82TWIKK355 pKa = 10.48DD356 pKa = 2.88WSAQIIGG363 pKa = 3.71
MM1 pKa = 6.95NHH3 pKa = 5.75HH4 pKa = 6.59TATHH8 pKa = 5.32TQATRR13 pKa = 11.84RR14 pKa = 11.84PLTTTLTALTLTAALAGCVPGSSNEE39 pKa = 3.98DD40 pKa = 3.24NAQGEE45 pKa = 4.74TPSTVTVVVHH55 pKa = 7.28DD56 pKa = 4.16SFQMSEE62 pKa = 3.93EE63 pKa = 3.93QQADD67 pKa = 3.73FEE69 pKa = 4.63KK70 pKa = 10.55TSGYY74 pKa = 11.06DD75 pKa = 3.2LTIVTNGDD83 pKa = 2.76GGALVNSLILTKK95 pKa = 10.44DD96 pKa = 3.19APIADD101 pKa = 3.46AVFGVDD107 pKa = 2.9NTFASRR113 pKa = 11.84GISEE117 pKa = 4.79EE118 pKa = 3.79VFAPYY123 pKa = 9.74TSEE126 pKa = 4.13VASQTSRR133 pKa = 11.84DD134 pKa = 3.49LALPGDD140 pKa = 4.23DD141 pKa = 4.09TLTAIDD147 pKa = 4.99FGDD150 pKa = 3.29VCLNVDD156 pKa = 3.59PTWFEE161 pKa = 3.8NHH163 pKa = 6.31SLDD166 pKa = 5.04VPATLDD172 pKa = 3.75DD173 pKa = 4.67LADD176 pKa = 3.7PAYY179 pKa = 10.88RR180 pKa = 11.84NLTVVTNPATSSPGLSFLFATIGKK204 pKa = 9.13YY205 pKa = 10.71GEE207 pKa = 5.08DD208 pKa = 3.88GYY210 pKa = 11.56LDD212 pKa = 3.67YY213 pKa = 8.34WTQLKK218 pKa = 11.03DD219 pKa = 3.35NGLKK223 pKa = 9.97IVDD226 pKa = 3.86GWEE229 pKa = 3.39DD230 pKa = 3.24AYY232 pKa = 11.66YY233 pKa = 11.24VDD235 pKa = 5.62FTATGDD241 pKa = 3.5GDD243 pKa = 3.91RR244 pKa = 11.84PIALSYY250 pKa = 10.85ASSPAFTVTDD260 pKa = 4.39DD261 pKa = 3.96GSATTTSALLDD272 pKa = 3.36TCFRR276 pKa = 11.84QVEE279 pKa = 4.46YY280 pKa = 11.15AGVLHH285 pKa = 6.87GAANPEE291 pKa = 3.98GAQAFIDD298 pKa = 3.87YY299 pKa = 10.22LLSEE303 pKa = 5.57DD304 pKa = 4.22YY305 pKa = 9.65QTTIPDD311 pKa = 3.58AMYY314 pKa = 8.45MYY316 pKa = 10.21PVADD320 pKa = 3.61TVEE323 pKa = 5.4LPDD326 pKa = 3.69AWAQFAPVPTTPISLDD342 pKa = 3.33PDD344 pKa = 4.21TIARR348 pKa = 11.84MRR350 pKa = 11.84DD351 pKa = 2.82TWIKK355 pKa = 10.48DD356 pKa = 2.88WSAQIIGG363 pKa = 3.71
Molecular weight: 38.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7R573|C7R573_JONDD Regulatory protein ArsR OS=Jonesia denitrificans (strain ATCC 14870 / DSM 20603 / BCRC 15368 / CIP 55.134 / JCM 11481 / NBRC 15587 / NCTC 10816 / Prevot 55134) OX=471856 GN=Jden_0075 PE=4 SV=1
MM1 pKa = 7.65KK2 pKa = 10.25VRR4 pKa = 11.84NSLKK8 pKa = 10.4SLKK11 pKa = 9.52NQPGSQIVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84VFVINKK30 pKa = 9.17KK31 pKa = 9.28NPRR34 pKa = 11.84LKK36 pKa = 10.61ARR38 pKa = 11.84QGG40 pKa = 3.23
MM1 pKa = 7.65KK2 pKa = 10.25VRR4 pKa = 11.84NSLKK8 pKa = 10.4SLKK11 pKa = 9.52NQPGSQIVRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84GRR24 pKa = 11.84VFVINKK30 pKa = 9.17KK31 pKa = 9.28NPRR34 pKa = 11.84LKK36 pKa = 10.61ARR38 pKa = 11.84QGG40 pKa = 3.23
Molecular weight: 4.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
819653 |
30 |
2043 |
328.5 |
35.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.756 ± 0.062 | 0.631 ± 0.012 |
6.264 ± 0.044 | 5.214 ± 0.046 |
2.941 ± 0.028 | 8.16 ± 0.046 |
2.709 ± 0.034 | 4.826 ± 0.031 |
2.263 ± 0.035 | 9.484 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.019 ± 0.018 | 2.544 ± 0.032 |
5.372 ± 0.04 | 3.44 ± 0.026 |
6.522 ± 0.05 | 5.945 ± 0.04 |
7.425 ± 0.064 | 8.882 ± 0.059 |
1.476 ± 0.021 | 2.124 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
