
Human papillomavirus type 203
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; unclassified Papillomaviridae; Human papillomavirus types
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
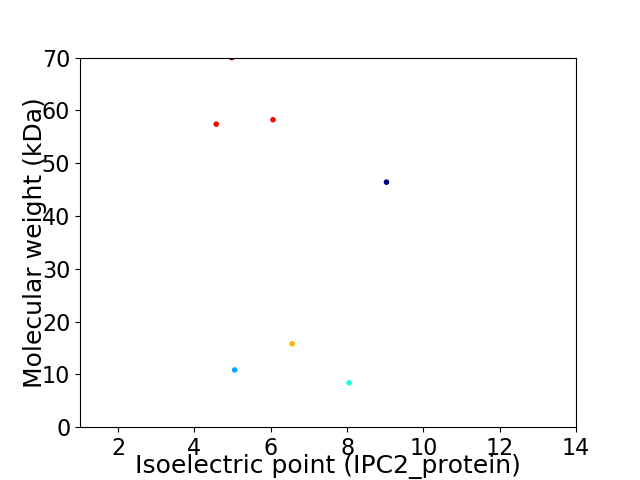
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2L1K401|A0A2L1K401_9PAPI Regulatory protein E2 OS=Human papillomavirus type 203 OX=2093789 GN=E2 PE=3 SV=1
MM1 pKa = 7.42SLTRR5 pKa = 11.84ARR7 pKa = 11.84RR8 pKa = 11.84VKK10 pKa = 10.33RR11 pKa = 11.84DD12 pKa = 3.33SVQNLYY18 pKa = 9.71NQCQLTGNCLPDD30 pKa = 3.28VKK32 pKa = 10.99NKK34 pKa = 10.04VEE36 pKa = 4.14GTTLADD42 pKa = 3.35KK43 pKa = 10.52LLKK46 pKa = 10.12IFSSIIYY53 pKa = 10.23LGGLSIGTGKK63 pKa = 8.72GTGGSTGYY71 pKa = 8.83TPLGGTTGGGRR82 pKa = 11.84VGTGGTVVRR91 pKa = 11.84PNVVVEE97 pKa = 4.04PLGPTDD103 pKa = 3.78IVPVDD108 pKa = 3.64TLNPNSSSIVPLLEE122 pKa = 4.59AGPDD126 pKa = 3.6IIISEE131 pKa = 4.47SAPGPKK137 pKa = 9.84VNLPDD142 pKa = 4.43AGTDD146 pKa = 3.52LGISEE151 pKa = 5.24LDD153 pKa = 3.04IVTTSDD159 pKa = 3.29PVSDD163 pKa = 3.75VTIGNAGGPTVSTSEE178 pKa = 4.24DD179 pKa = 3.22SVAIIDD185 pKa = 3.94VQPASSTPRR194 pKa = 11.84RR195 pKa = 11.84VATSTYY201 pKa = 10.16NNPSYY206 pKa = 9.93VTVYY210 pKa = 10.4SQSTLAEE217 pKa = 3.98EE218 pKa = 4.25TSANINVFVDD228 pKa = 3.51SAFGGDD234 pKa = 3.78VIGSSGQDD242 pKa = 2.81IPLEE246 pKa = 4.19TFNVSSDD253 pKa = 3.29LAEE256 pKa = 4.34FEE258 pKa = 4.45IEE260 pKa = 4.14EE261 pKa = 4.52PPRR264 pKa = 11.84TSTPRR269 pKa = 11.84DD270 pKa = 3.42TIEE273 pKa = 4.5RR274 pKa = 11.84ALQRR278 pKa = 11.84ARR280 pKa = 11.84DD281 pKa = 3.89LYY283 pKa = 9.81QRR285 pKa = 11.84RR286 pKa = 11.84VQQVRR291 pKa = 11.84TRR293 pKa = 11.84NVDD296 pKa = 3.73FLGRR300 pKa = 11.84PQMAVQFQFEE310 pKa = 4.32NPAFEE315 pKa = 5.95DD316 pKa = 4.96DD317 pKa = 3.2ITLTFEE323 pKa = 4.07QDD325 pKa = 3.24LNQLATAAPDD335 pKa = 3.56ADD337 pKa = 3.77FADD340 pKa = 4.92VIRR343 pKa = 11.84LHH345 pKa = 6.5RR346 pKa = 11.84PRR348 pKa = 11.84YY349 pKa = 8.97SVTDD353 pKa = 3.44AGTVRR358 pKa = 11.84VSRR361 pKa = 11.84LGQRR365 pKa = 11.84GTIHH369 pKa = 6.15TRR371 pKa = 11.84SGLQIGQQVHH381 pKa = 6.5FYY383 pKa = 11.02YY384 pKa = 10.49DD385 pKa = 2.97ISDD388 pKa = 3.89IEE390 pKa = 3.97PAEE393 pKa = 4.48AIEE396 pKa = 3.96LTLLGEE402 pKa = 4.58HH403 pKa = 6.48SGEE406 pKa = 3.88AAVVNAQSEE415 pKa = 4.91STVVDD420 pKa = 3.79AEE422 pKa = 4.35NSNSPFLYY430 pKa = 10.1PEE432 pKa = 4.42EE433 pKa = 4.13EE434 pKa = 4.97LIDD437 pKa = 3.88VPTEE441 pKa = 4.1DD442 pKa = 4.98FSDD445 pKa = 3.46SHH447 pKa = 7.57LVLSSNSRR455 pKa = 11.84RR456 pKa = 11.84SYY458 pKa = 9.21ITTPTLPPGTALRR471 pKa = 11.84VFIDD475 pKa = 4.49DD476 pKa = 3.73YY477 pKa = 11.94ANGLFVSYY485 pKa = 10.42PEE487 pKa = 4.78TYY489 pKa = 10.31NPPQIIVPAEE499 pKa = 3.9NLPPTVLIDD508 pKa = 3.67SFNSNDD514 pKa = 3.97FILHH518 pKa = 6.23PSHH521 pKa = 6.74NKK523 pKa = 8.03RR524 pKa = 11.84RR525 pKa = 11.84KK526 pKa = 9.04RR527 pKa = 11.84KK528 pKa = 9.99RR529 pKa = 11.84GDD531 pKa = 3.05VV532 pKa = 3.33
MM1 pKa = 7.42SLTRR5 pKa = 11.84ARR7 pKa = 11.84RR8 pKa = 11.84VKK10 pKa = 10.33RR11 pKa = 11.84DD12 pKa = 3.33SVQNLYY18 pKa = 9.71NQCQLTGNCLPDD30 pKa = 3.28VKK32 pKa = 10.99NKK34 pKa = 10.04VEE36 pKa = 4.14GTTLADD42 pKa = 3.35KK43 pKa = 10.52LLKK46 pKa = 10.12IFSSIIYY53 pKa = 10.23LGGLSIGTGKK63 pKa = 8.72GTGGSTGYY71 pKa = 8.83TPLGGTTGGGRR82 pKa = 11.84VGTGGTVVRR91 pKa = 11.84PNVVVEE97 pKa = 4.04PLGPTDD103 pKa = 3.78IVPVDD108 pKa = 3.64TLNPNSSSIVPLLEE122 pKa = 4.59AGPDD126 pKa = 3.6IIISEE131 pKa = 4.47SAPGPKK137 pKa = 9.84VNLPDD142 pKa = 4.43AGTDD146 pKa = 3.52LGISEE151 pKa = 5.24LDD153 pKa = 3.04IVTTSDD159 pKa = 3.29PVSDD163 pKa = 3.75VTIGNAGGPTVSTSEE178 pKa = 4.24DD179 pKa = 3.22SVAIIDD185 pKa = 3.94VQPASSTPRR194 pKa = 11.84RR195 pKa = 11.84VATSTYY201 pKa = 10.16NNPSYY206 pKa = 9.93VTVYY210 pKa = 10.4SQSTLAEE217 pKa = 3.98EE218 pKa = 4.25TSANINVFVDD228 pKa = 3.51SAFGGDD234 pKa = 3.78VIGSSGQDD242 pKa = 2.81IPLEE246 pKa = 4.19TFNVSSDD253 pKa = 3.29LAEE256 pKa = 4.34FEE258 pKa = 4.45IEE260 pKa = 4.14EE261 pKa = 4.52PPRR264 pKa = 11.84TSTPRR269 pKa = 11.84DD270 pKa = 3.42TIEE273 pKa = 4.5RR274 pKa = 11.84ALQRR278 pKa = 11.84ARR280 pKa = 11.84DD281 pKa = 3.89LYY283 pKa = 9.81QRR285 pKa = 11.84RR286 pKa = 11.84VQQVRR291 pKa = 11.84TRR293 pKa = 11.84NVDD296 pKa = 3.73FLGRR300 pKa = 11.84PQMAVQFQFEE310 pKa = 4.32NPAFEE315 pKa = 5.95DD316 pKa = 4.96DD317 pKa = 3.2ITLTFEE323 pKa = 4.07QDD325 pKa = 3.24LNQLATAAPDD335 pKa = 3.56ADD337 pKa = 3.77FADD340 pKa = 4.92VIRR343 pKa = 11.84LHH345 pKa = 6.5RR346 pKa = 11.84PRR348 pKa = 11.84YY349 pKa = 8.97SVTDD353 pKa = 3.44AGTVRR358 pKa = 11.84VSRR361 pKa = 11.84LGQRR365 pKa = 11.84GTIHH369 pKa = 6.15TRR371 pKa = 11.84SGLQIGQQVHH381 pKa = 6.5FYY383 pKa = 11.02YY384 pKa = 10.49DD385 pKa = 2.97ISDD388 pKa = 3.89IEE390 pKa = 3.97PAEE393 pKa = 4.48AIEE396 pKa = 3.96LTLLGEE402 pKa = 4.58HH403 pKa = 6.48SGEE406 pKa = 3.88AAVVNAQSEE415 pKa = 4.91STVVDD420 pKa = 3.79AEE422 pKa = 4.35NSNSPFLYY430 pKa = 10.1PEE432 pKa = 4.42EE433 pKa = 4.13EE434 pKa = 4.97LIDD437 pKa = 3.88VPTEE441 pKa = 4.1DD442 pKa = 4.98FSDD445 pKa = 3.46SHH447 pKa = 7.57LVLSSNSRR455 pKa = 11.84RR456 pKa = 11.84SYY458 pKa = 9.21ITTPTLPPGTALRR471 pKa = 11.84VFIDD475 pKa = 4.49DD476 pKa = 3.73YY477 pKa = 11.94ANGLFVSYY485 pKa = 10.42PEE487 pKa = 4.78TYY489 pKa = 10.31NPPQIIVPAEE499 pKa = 3.9NLPPTVLIDD508 pKa = 3.67SFNSNDD514 pKa = 3.97FILHH518 pKa = 6.23PSHH521 pKa = 6.74NKK523 pKa = 8.03RR524 pKa = 11.84RR525 pKa = 11.84KK526 pKa = 9.04RR527 pKa = 11.84KK528 pKa = 9.99RR529 pKa = 11.84GDD531 pKa = 3.05VV532 pKa = 3.33
Molecular weight: 57.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2L1K403|A0A2L1K403_9PAPI Major capsid protein L1 OS=Human papillomavirus type 203 OX=2093789 GN=L1 PE=3 SV=1
MM1 pKa = 7.47EE2 pKa = 4.9SLRR5 pKa = 11.84EE6 pKa = 3.93RR7 pKa = 11.84FDD9 pKa = 3.65ALQEE13 pKa = 3.98KK14 pKa = 10.4LIEE17 pKa = 4.36IYY19 pKa = 10.4EE20 pKa = 4.38AGSTNLEE27 pKa = 4.07DD28 pKa = 5.48QIKK31 pKa = 9.79HH32 pKa = 4.58WDD34 pKa = 3.28IVRR37 pKa = 11.84KK38 pKa = 9.69EE39 pKa = 3.95SVIMHH44 pKa = 6.28FARR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 8.8GLTRR53 pKa = 11.84LGLQQLPALTVSEE66 pKa = 4.63TNAKK70 pKa = 8.7NAIMMSIYY78 pKa = 10.31LQSLNNSQYY87 pKa = 11.49ASEE90 pKa = 4.18SWGLSDD96 pKa = 4.41TSLEE100 pKa = 4.32LFLTAPMHH108 pKa = 5.26TFKK111 pKa = 11.05KK112 pKa = 9.76SGYY115 pKa = 8.92SVDD118 pKa = 3.62VLFDD122 pKa = 3.73NDD124 pKa = 3.36EE125 pKa = 4.77DD126 pKa = 3.96NLYY129 pKa = 10.19TYY131 pKa = 7.36TAWRR135 pKa = 11.84FVYY138 pKa = 10.55YY139 pKa = 10.28QDD141 pKa = 6.3DD142 pKa = 3.56EE143 pKa = 5.28GEE145 pKa = 4.04WHH147 pKa = 6.29KK148 pKa = 11.39VEE150 pKa = 5.48SGVDD154 pKa = 3.41YY155 pKa = 11.37DD156 pKa = 4.33GIFYY160 pKa = 8.73KK161 pKa = 10.38TVEE164 pKa = 4.22NHH166 pKa = 4.14TEE168 pKa = 3.84YY169 pKa = 11.11YY170 pKa = 10.11IRR172 pKa = 11.84FQDD175 pKa = 3.66DD176 pKa = 3.25APRR179 pKa = 11.84FSRR182 pKa = 11.84TGRR185 pKa = 11.84WTVKK189 pKa = 10.44YY190 pKa = 9.94KK191 pKa = 10.76SQTISTSVTSTSGSDD206 pKa = 3.26PVSSSEE212 pKa = 4.9RR213 pKa = 11.84PTQQSSDD220 pKa = 3.7TLDD223 pKa = 4.39APPQPKK229 pKa = 9.7KK230 pKa = 10.71FRR232 pKa = 11.84VRR234 pKa = 11.84PPQQTGPSRR243 pKa = 11.84VSSSSNSKK251 pKa = 5.43TTKK254 pKa = 10.17KK255 pKa = 10.25EE256 pKa = 4.05SPKK259 pKa = 9.01TRR261 pKa = 11.84LRR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84RR266 pKa = 11.84GEE268 pKa = 4.09GEE270 pKa = 3.66PTTRR274 pKa = 11.84ATNRR278 pKa = 11.84RR279 pKa = 11.84TTSGRR284 pKa = 11.84ARR286 pKa = 11.84RR287 pKa = 11.84RR288 pKa = 11.84LEE290 pKa = 3.66YY291 pKa = 10.16AYY293 pKa = 7.06PTPEE297 pKa = 3.77QVGSVHH303 pKa = 7.48RR304 pKa = 11.84LPQGSNLGRR313 pKa = 11.84LGRR316 pKa = 11.84LQEE319 pKa = 4.12EE320 pKa = 4.38ARR322 pKa = 11.84DD323 pKa = 3.85PPVVLLRR330 pKa = 11.84GLANTLKK337 pKa = 10.55CFRR340 pKa = 11.84FRR342 pKa = 11.84CKK344 pKa = 10.02QKK346 pKa = 10.74YY347 pKa = 9.45KK348 pKa = 11.17GLFQHH353 pKa = 7.24ISTTFSWVGEE363 pKa = 4.27GYY365 pKa = 10.31EE366 pKa = 3.93RR367 pKa = 11.84LGNARR372 pKa = 11.84ILIAFQSTAQRR383 pKa = 11.84EE384 pKa = 4.25KK385 pKa = 10.03FLAVAVLPKK394 pKa = 9.71GTEE397 pKa = 3.89VVYY400 pKa = 10.99GSLNALL406 pKa = 3.72
MM1 pKa = 7.47EE2 pKa = 4.9SLRR5 pKa = 11.84EE6 pKa = 3.93RR7 pKa = 11.84FDD9 pKa = 3.65ALQEE13 pKa = 3.98KK14 pKa = 10.4LIEE17 pKa = 4.36IYY19 pKa = 10.4EE20 pKa = 4.38AGSTNLEE27 pKa = 4.07DD28 pKa = 5.48QIKK31 pKa = 9.79HH32 pKa = 4.58WDD34 pKa = 3.28IVRR37 pKa = 11.84KK38 pKa = 9.69EE39 pKa = 3.95SVIMHH44 pKa = 6.28FARR47 pKa = 11.84RR48 pKa = 11.84KK49 pKa = 8.8GLTRR53 pKa = 11.84LGLQQLPALTVSEE66 pKa = 4.63TNAKK70 pKa = 8.7NAIMMSIYY78 pKa = 10.31LQSLNNSQYY87 pKa = 11.49ASEE90 pKa = 4.18SWGLSDD96 pKa = 4.41TSLEE100 pKa = 4.32LFLTAPMHH108 pKa = 5.26TFKK111 pKa = 11.05KK112 pKa = 9.76SGYY115 pKa = 8.92SVDD118 pKa = 3.62VLFDD122 pKa = 3.73NDD124 pKa = 3.36EE125 pKa = 4.77DD126 pKa = 3.96NLYY129 pKa = 10.19TYY131 pKa = 7.36TAWRR135 pKa = 11.84FVYY138 pKa = 10.55YY139 pKa = 10.28QDD141 pKa = 6.3DD142 pKa = 3.56EE143 pKa = 5.28GEE145 pKa = 4.04WHH147 pKa = 6.29KK148 pKa = 11.39VEE150 pKa = 5.48SGVDD154 pKa = 3.41YY155 pKa = 11.37DD156 pKa = 4.33GIFYY160 pKa = 8.73KK161 pKa = 10.38TVEE164 pKa = 4.22NHH166 pKa = 4.14TEE168 pKa = 3.84YY169 pKa = 11.11YY170 pKa = 10.11IRR172 pKa = 11.84FQDD175 pKa = 3.66DD176 pKa = 3.25APRR179 pKa = 11.84FSRR182 pKa = 11.84TGRR185 pKa = 11.84WTVKK189 pKa = 10.44YY190 pKa = 9.94KK191 pKa = 10.76SQTISTSVTSTSGSDD206 pKa = 3.26PVSSSEE212 pKa = 4.9RR213 pKa = 11.84PTQQSSDD220 pKa = 3.7TLDD223 pKa = 4.39APPQPKK229 pKa = 9.7KK230 pKa = 10.71FRR232 pKa = 11.84VRR234 pKa = 11.84PPQQTGPSRR243 pKa = 11.84VSSSSNSKK251 pKa = 5.43TTKK254 pKa = 10.17KK255 pKa = 10.25EE256 pKa = 4.05SPKK259 pKa = 9.01TRR261 pKa = 11.84LRR263 pKa = 11.84RR264 pKa = 11.84RR265 pKa = 11.84RR266 pKa = 11.84GEE268 pKa = 4.09GEE270 pKa = 3.66PTTRR274 pKa = 11.84ATNRR278 pKa = 11.84RR279 pKa = 11.84TTSGRR284 pKa = 11.84ARR286 pKa = 11.84RR287 pKa = 11.84RR288 pKa = 11.84LEE290 pKa = 3.66YY291 pKa = 10.16AYY293 pKa = 7.06PTPEE297 pKa = 3.77QVGSVHH303 pKa = 7.48RR304 pKa = 11.84LPQGSNLGRR313 pKa = 11.84LGRR316 pKa = 11.84LQEE319 pKa = 4.12EE320 pKa = 4.38ARR322 pKa = 11.84DD323 pKa = 3.85PPVVLLRR330 pKa = 11.84GLANTLKK337 pKa = 10.55CFRR340 pKa = 11.84FRR342 pKa = 11.84CKK344 pKa = 10.02QKK346 pKa = 10.74YY347 pKa = 9.45KK348 pKa = 11.17GLFQHH353 pKa = 7.24ISTTFSWVGEE363 pKa = 4.27GYY365 pKa = 10.31EE366 pKa = 3.93RR367 pKa = 11.84LGNARR372 pKa = 11.84ILIAFQSTAQRR383 pKa = 11.84EE384 pKa = 4.25KK385 pKa = 10.03FLAVAVLPKK394 pKa = 9.71GTEE397 pKa = 3.89VVYY400 pKa = 10.99GSLNALL406 pKa = 3.72
Molecular weight: 46.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2370 |
70 |
613 |
338.6 |
38.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.485 ± 0.185 | 2.236 ± 1.004 |
6.371 ± 0.406 | 6.287 ± 0.64 |
4.388 ± 0.418 | 5.654 ± 0.644 |
1.772 ± 0.274 | 4.895 ± 0.653 |
5.021 ± 0.99 | 9.536 ± 0.79 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.857 ± 0.574 | 5.232 ± 0.683 |
5.485 ± 0.85 | 4.051 ± 0.344 |
5.907 ± 0.903 | 7.764 ± 0.693 |
6.54 ± 0.812 | 6.962 ± 0.596 |
1.181 ± 0.339 | 3.376 ± 0.307 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
