
Wheat dwarf virus (isolate Sweden) (WDV)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus; Wheat dwarf virus
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
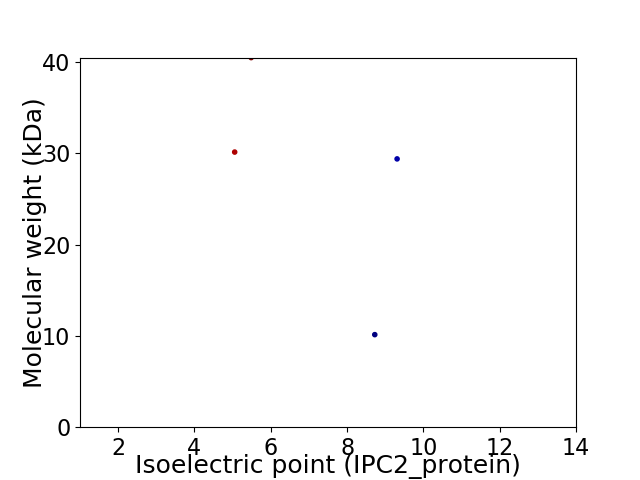
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
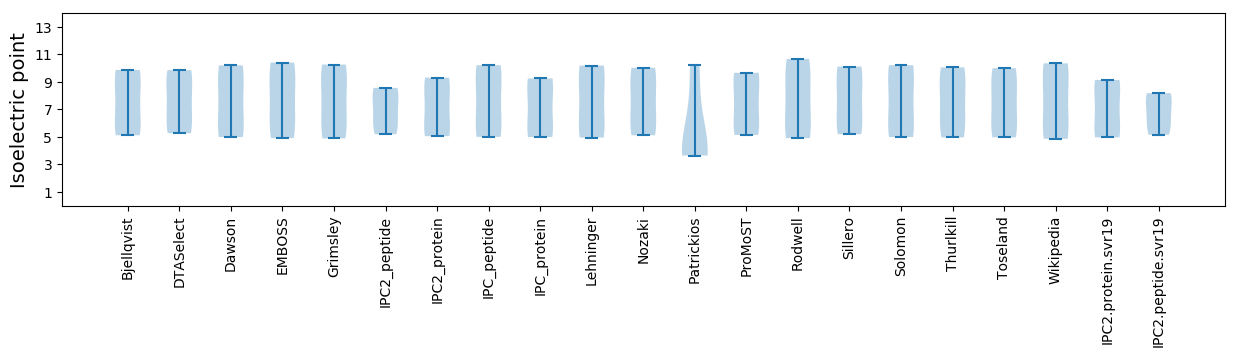
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P06849|MP_WDVS Movement protein OS=Wheat dwarf virus (isolate Sweden) OX=268789 GN=V2 PE=3 SV=1
MM1 pKa = 7.66ASSSAPRR8 pKa = 11.84FRR10 pKa = 11.84VYY12 pKa = 10.78SKK14 pKa = 11.3YY15 pKa = 11.01LFLTYY20 pKa = 9.26PEE22 pKa = 4.63CTLEE26 pKa = 3.85PQYY29 pKa = 11.83ALDD32 pKa = 4.26SLRR35 pKa = 11.84TLLNKK40 pKa = 10.04YY41 pKa = 9.36EE42 pKa = 4.24PLYY45 pKa = 10.1IAAVRR50 pKa = 11.84EE51 pKa = 4.02LHH53 pKa = 6.85EE54 pKa = 5.61DD55 pKa = 4.13GSPHH59 pKa = 6.69LHH61 pKa = 5.96VLVQNKK67 pKa = 8.63LRR69 pKa = 11.84ASITNPNALNLRR81 pKa = 11.84MDD83 pKa = 4.13TSPFSIFHH91 pKa = 6.53PNIQAAKK98 pKa = 9.81DD99 pKa = 3.72CNQVRR104 pKa = 11.84DD105 pKa = 4.27YY106 pKa = 9.75ITKK109 pKa = 10.41EE110 pKa = 3.51VDD112 pKa = 3.0SDD114 pKa = 4.11VNTAEE119 pKa = 3.73WGTFVAVSTPGRR131 pKa = 11.84KK132 pKa = 9.51DD133 pKa = 2.79RR134 pKa = 11.84DD135 pKa = 3.22ADD137 pKa = 3.4MKK139 pKa = 10.91QIIEE143 pKa = 4.28SSSSRR148 pKa = 11.84EE149 pKa = 3.73EE150 pKa = 4.01FLSMVCNRR158 pKa = 11.84FPFEE162 pKa = 3.45WSIRR166 pKa = 11.84LKK168 pKa = 10.83DD169 pKa = 3.77FEE171 pKa = 4.34YY172 pKa = 10.01TARR175 pKa = 11.84HH176 pKa = 6.38LFPDD180 pKa = 4.45PVATYY185 pKa = 9.18TPEE188 pKa = 4.05FPTEE192 pKa = 3.81SLICHH197 pKa = 5.52EE198 pKa = 5.27TIEE201 pKa = 4.35SWKK204 pKa = 10.62NEE206 pKa = 3.76HH207 pKa = 6.69LYY209 pKa = 10.78SVSLEE214 pKa = 4.09SYY216 pKa = 9.72ILCTSTPADD225 pKa = 3.44QAQSDD230 pKa = 4.74LEE232 pKa = 4.23WMDD235 pKa = 4.06DD236 pKa = 3.49YY237 pKa = 11.8SRR239 pKa = 11.84SHH241 pKa = 7.0RR242 pKa = 11.84GGISPSTSAGQPEE255 pKa = 4.43QEE257 pKa = 4.2RR258 pKa = 11.84LPGQGLL264 pKa = 3.44
MM1 pKa = 7.66ASSSAPRR8 pKa = 11.84FRR10 pKa = 11.84VYY12 pKa = 10.78SKK14 pKa = 11.3YY15 pKa = 11.01LFLTYY20 pKa = 9.26PEE22 pKa = 4.63CTLEE26 pKa = 3.85PQYY29 pKa = 11.83ALDD32 pKa = 4.26SLRR35 pKa = 11.84TLLNKK40 pKa = 10.04YY41 pKa = 9.36EE42 pKa = 4.24PLYY45 pKa = 10.1IAAVRR50 pKa = 11.84EE51 pKa = 4.02LHH53 pKa = 6.85EE54 pKa = 5.61DD55 pKa = 4.13GSPHH59 pKa = 6.69LHH61 pKa = 5.96VLVQNKK67 pKa = 8.63LRR69 pKa = 11.84ASITNPNALNLRR81 pKa = 11.84MDD83 pKa = 4.13TSPFSIFHH91 pKa = 6.53PNIQAAKK98 pKa = 9.81DD99 pKa = 3.72CNQVRR104 pKa = 11.84DD105 pKa = 4.27YY106 pKa = 9.75ITKK109 pKa = 10.41EE110 pKa = 3.51VDD112 pKa = 3.0SDD114 pKa = 4.11VNTAEE119 pKa = 3.73WGTFVAVSTPGRR131 pKa = 11.84KK132 pKa = 9.51DD133 pKa = 2.79RR134 pKa = 11.84DD135 pKa = 3.22ADD137 pKa = 3.4MKK139 pKa = 10.91QIIEE143 pKa = 4.28SSSSRR148 pKa = 11.84EE149 pKa = 3.73EE150 pKa = 4.01FLSMVCNRR158 pKa = 11.84FPFEE162 pKa = 3.45WSIRR166 pKa = 11.84LKK168 pKa = 10.83DD169 pKa = 3.77FEE171 pKa = 4.34YY172 pKa = 10.01TARR175 pKa = 11.84HH176 pKa = 6.38LFPDD180 pKa = 4.45PVATYY185 pKa = 9.18TPEE188 pKa = 4.05FPTEE192 pKa = 3.81SLICHH197 pKa = 5.52EE198 pKa = 5.27TIEE201 pKa = 4.35SWKK204 pKa = 10.62NEE206 pKa = 3.76HH207 pKa = 6.69LYY209 pKa = 10.78SVSLEE214 pKa = 4.09SYY216 pKa = 9.72ILCTSTPADD225 pKa = 3.44QAQSDD230 pKa = 4.74LEE232 pKa = 4.23WMDD235 pKa = 4.06DD236 pKa = 3.49YY237 pKa = 11.8SRR239 pKa = 11.84SHH241 pKa = 7.0RR242 pKa = 11.84GGISPSTSAGQPEE255 pKa = 4.43QEE257 pKa = 4.2RR258 pKa = 11.84LPGQGLL264 pKa = 3.44
Molecular weight: 30.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q67622|REP_WDVS Replication-associated protein OS=Wheat dwarf virus (isolate Sweden) OX=268789 GN=C1/C2 PE=1 SV=2
MM1 pKa = 6.79VTNKK5 pKa = 10.32DD6 pKa = 3.24SRR8 pKa = 11.84GKK10 pKa = 10.42GKK12 pKa = 10.62RR13 pKa = 11.84KK14 pKa = 8.95MEE16 pKa = 4.43EE17 pKa = 4.38GEE19 pKa = 4.34SSGRR23 pKa = 11.84WKK25 pKa = 10.56GAVYY29 pKa = 10.24KK30 pKa = 10.38RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 7.79QAYY36 pKa = 8.07KK37 pKa = 10.14VVPVKK42 pKa = 10.61PPALCVFRR50 pKa = 11.84YY51 pKa = 9.52NWLNSDD57 pKa = 3.32RR58 pKa = 11.84TNIVVGNTPRR68 pKa = 11.84VDD70 pKa = 5.54LITCFAQGKK79 pKa = 9.49ADD81 pKa = 3.94NNRR84 pKa = 11.84HH85 pKa = 4.81TNQTVLYY92 pKa = 8.93KK93 pKa = 10.21FNIQGTCYY101 pKa = 9.88MSDD104 pKa = 2.84ASAPFIGPVRR114 pKa = 11.84LYY116 pKa = 10.41HH117 pKa = 5.83WLVYY121 pKa = 10.28DD122 pKa = 4.74AEE124 pKa = 4.87PKK126 pKa = 10.12QAMPDD131 pKa = 3.21ATDD134 pKa = 3.14IFTMPWNLLPSTWTVQRR151 pKa = 11.84AWSHH155 pKa = 5.48RR156 pKa = 11.84FVVKK160 pKa = 10.5RR161 pKa = 11.84KK162 pKa = 5.39WTVNLVTDD170 pKa = 4.01GRR172 pKa = 11.84KK173 pKa = 9.77VGSKK177 pKa = 9.13TVDD180 pKa = 2.98QRR182 pKa = 11.84YY183 pKa = 8.54NWVVGKK189 pKa = 10.44NIVDD193 pKa = 3.38ANKK196 pKa = 9.44FFKK199 pKa = 10.54GLRR202 pKa = 11.84VTTEE206 pKa = 3.21WMNTGDD212 pKa = 5.0GKK214 pKa = 10.85IGDD217 pKa = 3.88IKK219 pKa = 10.84KK220 pKa = 10.23GALYY224 pKa = 10.24LISSTRR230 pKa = 11.84GGVTGDD236 pKa = 3.47SASTAFDD243 pKa = 3.61VVCAYY248 pKa = 8.43THH250 pKa = 6.31ACYY253 pKa = 10.13FKK255 pKa = 10.94AIGIQQ260 pKa = 3.17
MM1 pKa = 6.79VTNKK5 pKa = 10.32DD6 pKa = 3.24SRR8 pKa = 11.84GKK10 pKa = 10.42GKK12 pKa = 10.62RR13 pKa = 11.84KK14 pKa = 8.95MEE16 pKa = 4.43EE17 pKa = 4.38GEE19 pKa = 4.34SSGRR23 pKa = 11.84WKK25 pKa = 10.56GAVYY29 pKa = 10.24KK30 pKa = 10.38RR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 7.79QAYY36 pKa = 8.07KK37 pKa = 10.14VVPVKK42 pKa = 10.61PPALCVFRR50 pKa = 11.84YY51 pKa = 9.52NWLNSDD57 pKa = 3.32RR58 pKa = 11.84TNIVVGNTPRR68 pKa = 11.84VDD70 pKa = 5.54LITCFAQGKK79 pKa = 9.49ADD81 pKa = 3.94NNRR84 pKa = 11.84HH85 pKa = 4.81TNQTVLYY92 pKa = 8.93KK93 pKa = 10.21FNIQGTCYY101 pKa = 9.88MSDD104 pKa = 2.84ASAPFIGPVRR114 pKa = 11.84LYY116 pKa = 10.41HH117 pKa = 5.83WLVYY121 pKa = 10.28DD122 pKa = 4.74AEE124 pKa = 4.87PKK126 pKa = 10.12QAMPDD131 pKa = 3.21ATDD134 pKa = 3.14IFTMPWNLLPSTWTVQRR151 pKa = 11.84AWSHH155 pKa = 5.48RR156 pKa = 11.84FVVKK160 pKa = 10.5RR161 pKa = 11.84KK162 pKa = 5.39WTVNLVTDD170 pKa = 4.01GRR172 pKa = 11.84KK173 pKa = 9.77VGSKK177 pKa = 9.13TVDD180 pKa = 2.98QRR182 pKa = 11.84YY183 pKa = 8.54NWVVGKK189 pKa = 10.44NIVDD193 pKa = 3.38ANKK196 pKa = 9.44FFKK199 pKa = 10.54GLRR202 pKa = 11.84VTTEE206 pKa = 3.21WMNTGDD212 pKa = 5.0GKK214 pKa = 10.85IGDD217 pKa = 3.88IKK219 pKa = 10.84KK220 pKa = 10.23GALYY224 pKa = 10.24LISSTRR230 pKa = 11.84GGVTGDD236 pKa = 3.47SASTAFDD243 pKa = 3.61VVCAYY248 pKa = 8.43THH250 pKa = 6.31ACYY253 pKa = 10.13FKK255 pKa = 10.94AIGIQQ260 pKa = 3.17
Molecular weight: 29.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
965 |
90 |
351 |
241.3 |
27.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.425 ± 0.477 | 1.865 ± 0.094 |
5.907 ± 0.212 | 5.389 ± 1.205 |
4.456 ± 0.313 | 5.078 ± 1.099 |
2.28 ± 0.426 | 5.078 ± 0.402 |
5.596 ± 1.009 | 7.15 ± 0.925 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.969 ± 0.124 | 4.767 ± 0.503 |
5.907 ± 0.606 | 3.316 ± 0.539 |
6.01 ± 0.272 | 7.772 ± 1.255 |
7.254 ± 0.309 | 6.528 ± 1.053 |
2.176 ± 0.402 | 5.078 ± 0.444 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
