
Mycobacterium asiaticum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia;
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
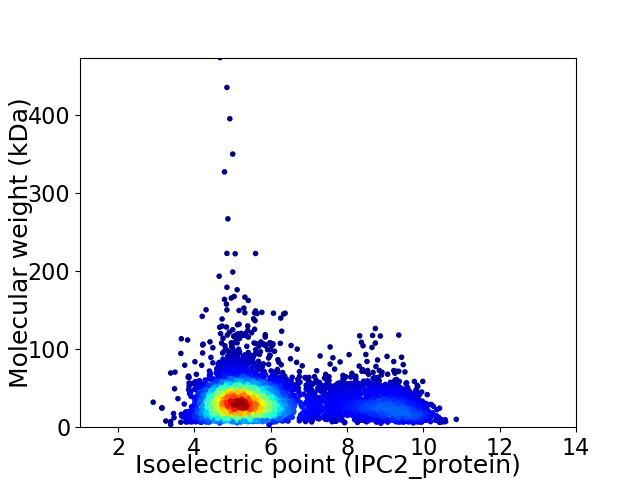
Virtual 2D-PAGE plot for 4763 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1A3N7G4|A0A1A3N7G4_MYCAS Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase OS=Mycobacterium asiaticum OX=1790 GN=cobT PE=3 SV=1
MM1 pKa = 7.76AAPDD5 pKa = 4.25LMTAAAANAAGIGSSIGAANLAALSPTSTVLAAAADD41 pKa = 4.07EE42 pKa = 4.43VSAAIASLFANHH54 pKa = 6.93ALDD57 pKa = 3.72YY58 pKa = 9.83QTFSAQVAGSYY69 pKa = 8.09EE70 pKa = 4.1QFVRR74 pKa = 11.84TLSAASGAYY83 pKa = 9.3AAAEE87 pKa = 4.03AASAGPLQPVLDD99 pKa = 4.67VINAPTNFLLGRR111 pKa = 11.84PLIGDD116 pKa = 4.29GVNGVAGTGQAGGDD130 pKa = 3.94GGILWGNGGTGGSGGAGQAGGRR152 pKa = 11.84GGNAGLIGDD161 pKa = 5.01GGTGGTGGLRR171 pKa = 11.84GGVGGAGGTGGWLLGNGGAGGTGGFGGNLYY201 pKa = 10.99GGGTIPAGAGGIGGRR216 pKa = 11.84ALSLFGASGATGNVGTFRR234 pKa = 11.84NVTFVSAEE242 pKa = 4.04DD243 pKa = 3.43AAVWAMANPDD253 pKa = 4.02ANFLLIGTDD262 pKa = 3.43GTNLSRR268 pKa = 11.84ILADD272 pKa = 3.78AAGTPNFHH280 pKa = 7.6ALMEE284 pKa = 4.13QSVTSASTVIGHH296 pKa = 5.39TTVSNPSWTTIQTGVWSEE314 pKa = 4.17TAGVTNNIFSPWTYY328 pKa = 11.28DD329 pKa = 3.62DD330 pKa = 3.74WPTVYY335 pKa = 10.8NHH337 pKa = 7.15LEE339 pKa = 3.71NAYY342 pKa = 10.22GDD344 pKa = 4.17GVNTTVIADD353 pKa = 3.58WKK355 pKa = 10.53VITDD359 pKa = 3.57IAGAGSNPADD369 pKa = 4.09NITFVPQVAGDD380 pKa = 4.21TNWEE384 pKa = 3.99ATSNLVAANAIDD396 pKa = 5.21AIAAADD402 pKa = 3.91PDD404 pKa = 3.79KK405 pKa = 11.63GNLIFAYY412 pKa = 8.5FTGPDD417 pKa = 3.89VNAHH421 pKa = 6.67AYY423 pKa = 10.31GGDD426 pKa = 3.67SPQYY430 pKa = 9.74AAALRR435 pKa = 11.84TMDD438 pKa = 4.77ANLGSQTNGTGMLGAVAQWEE458 pKa = 4.12AANPGEE464 pKa = 4.37EE465 pKa = 3.99FSVLVVTDD473 pKa = 3.73HH474 pKa = 7.12GEE476 pKa = 3.9LGPAQFGGITHH487 pKa = 7.33GFQSPLEE494 pKa = 4.14TASFVIFDD502 pKa = 3.62QAFDD506 pKa = 3.71NSSDD510 pKa = 3.5GLINNSWQIVSTTPTILDD528 pKa = 3.33QFYY531 pKa = 10.57LPQPSYY537 pKa = 10.16MQGAPITSPVFDD549 pKa = 3.94GTYY552 pKa = 10.52VDD554 pKa = 4.48PGPNLFSALSADD566 pKa = 4.79FAAQGYY572 pKa = 8.63PDD574 pKa = 3.67VVTNVSLSARR584 pKa = 11.84TLAASVPYY592 pKa = 10.38LAFEE596 pKa = 5.14PINGFIHH603 pKa = 6.99SMPEE607 pKa = 3.89FLQLPVSWIGAGVYY621 pKa = 7.41QTLNIPAQMFVRR633 pKa = 11.84FTGVTGNEE641 pKa = 4.28IIPPILNPFISS652 pKa = 3.51
MM1 pKa = 7.76AAPDD5 pKa = 4.25LMTAAAANAAGIGSSIGAANLAALSPTSTVLAAAADD41 pKa = 4.07EE42 pKa = 4.43VSAAIASLFANHH54 pKa = 6.93ALDD57 pKa = 3.72YY58 pKa = 9.83QTFSAQVAGSYY69 pKa = 8.09EE70 pKa = 4.1QFVRR74 pKa = 11.84TLSAASGAYY83 pKa = 9.3AAAEE87 pKa = 4.03AASAGPLQPVLDD99 pKa = 4.67VINAPTNFLLGRR111 pKa = 11.84PLIGDD116 pKa = 4.29GVNGVAGTGQAGGDD130 pKa = 3.94GGILWGNGGTGGSGGAGQAGGRR152 pKa = 11.84GGNAGLIGDD161 pKa = 5.01GGTGGTGGLRR171 pKa = 11.84GGVGGAGGTGGWLLGNGGAGGTGGFGGNLYY201 pKa = 10.99GGGTIPAGAGGIGGRR216 pKa = 11.84ALSLFGASGATGNVGTFRR234 pKa = 11.84NVTFVSAEE242 pKa = 4.04DD243 pKa = 3.43AAVWAMANPDD253 pKa = 4.02ANFLLIGTDD262 pKa = 3.43GTNLSRR268 pKa = 11.84ILADD272 pKa = 3.78AAGTPNFHH280 pKa = 7.6ALMEE284 pKa = 4.13QSVTSASTVIGHH296 pKa = 5.39TTVSNPSWTTIQTGVWSEE314 pKa = 4.17TAGVTNNIFSPWTYY328 pKa = 11.28DD329 pKa = 3.62DD330 pKa = 3.74WPTVYY335 pKa = 10.8NHH337 pKa = 7.15LEE339 pKa = 3.71NAYY342 pKa = 10.22GDD344 pKa = 4.17GVNTTVIADD353 pKa = 3.58WKK355 pKa = 10.53VITDD359 pKa = 3.57IAGAGSNPADD369 pKa = 4.09NITFVPQVAGDD380 pKa = 4.21TNWEE384 pKa = 3.99ATSNLVAANAIDD396 pKa = 5.21AIAAADD402 pKa = 3.91PDD404 pKa = 3.79KK405 pKa = 11.63GNLIFAYY412 pKa = 8.5FTGPDD417 pKa = 3.89VNAHH421 pKa = 6.67AYY423 pKa = 10.31GGDD426 pKa = 3.67SPQYY430 pKa = 9.74AAALRR435 pKa = 11.84TMDD438 pKa = 4.77ANLGSQTNGTGMLGAVAQWEE458 pKa = 4.12AANPGEE464 pKa = 4.37EE465 pKa = 3.99FSVLVVTDD473 pKa = 3.73HH474 pKa = 7.12GEE476 pKa = 3.9LGPAQFGGITHH487 pKa = 7.33GFQSPLEE494 pKa = 4.14TASFVIFDD502 pKa = 3.62QAFDD506 pKa = 3.71NSSDD510 pKa = 3.5GLINNSWQIVSTTPTILDD528 pKa = 3.33QFYY531 pKa = 10.57LPQPSYY537 pKa = 10.16MQGAPITSPVFDD549 pKa = 3.94GTYY552 pKa = 10.52VDD554 pKa = 4.48PGPNLFSALSADD566 pKa = 4.79FAAQGYY572 pKa = 8.63PDD574 pKa = 3.67VVTNVSLSARR584 pKa = 11.84TLAASVPYY592 pKa = 10.38LAFEE596 pKa = 5.14PINGFIHH603 pKa = 6.99SMPEE607 pKa = 3.89FLQLPVSWIGAGVYY621 pKa = 7.41QTLNIPAQMFVRR633 pKa = 11.84FTGVTGNEE641 pKa = 4.28IIPPILNPFISS652 pKa = 3.51
Molecular weight: 65.41 kDa
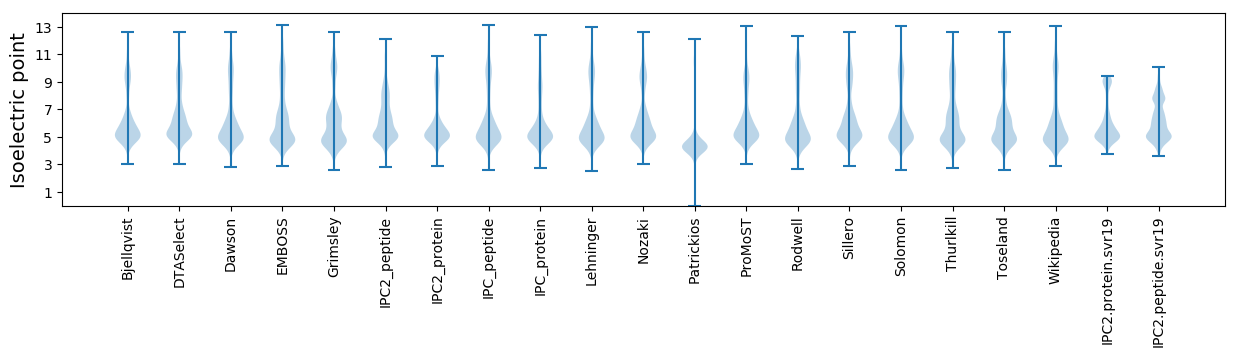
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1A3NE44|A0A1A3NE44_MYCAS Uncharacterized protein OS=Mycobacterium asiaticum OX=1790 GN=A5636_02035 PE=4 SV=1
MM1 pKa = 7.5QIVLLVAGAVFIISTLAVTVRR22 pKa = 11.84RR23 pKa = 11.84LMKK26 pKa = 10.32AAPHH30 pKa = 5.61SKK32 pKa = 10.04ARR34 pKa = 11.84AAMWGTLGQLGAVTIFLVLALQRR57 pKa = 11.84PHH59 pKa = 7.03SGNIFSWLILIALALAAINVVATLLIRR86 pKa = 11.84WLSSGIKK93 pKa = 9.37PP94 pKa = 4.05
MM1 pKa = 7.5QIVLLVAGAVFIISTLAVTVRR22 pKa = 11.84RR23 pKa = 11.84LMKK26 pKa = 10.32AAPHH30 pKa = 5.61SKK32 pKa = 10.04ARR34 pKa = 11.84AAMWGTLGQLGAVTIFLVLALQRR57 pKa = 11.84PHH59 pKa = 7.03SGNIFSWLILIALALAAINVVATLLIRR86 pKa = 11.84WLSSGIKK93 pKa = 9.37PP94 pKa = 4.05
Molecular weight: 10.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1590993 |
29 |
5956 |
334.0 |
35.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.823 ± 0.051 | 0.804 ± 0.01 |
6.106 ± 0.033 | 5.219 ± 0.036 |
3.067 ± 0.028 | 9.32 ± 0.18 |
2.219 ± 0.018 | 4.247 ± 0.028 |
2.218 ± 0.026 | 9.782 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.034 ± 0.017 | 2.481 ± 0.038 |
5.808 ± 0.034 | 3.183 ± 0.022 |
7.139 ± 0.046 | 5.532 ± 0.023 |
5.894 ± 0.021 | 8.454 ± 0.036 |
1.502 ± 0.015 | 2.166 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
