
Seonamhaeicola aphaedonensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Seonamhaeicola
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
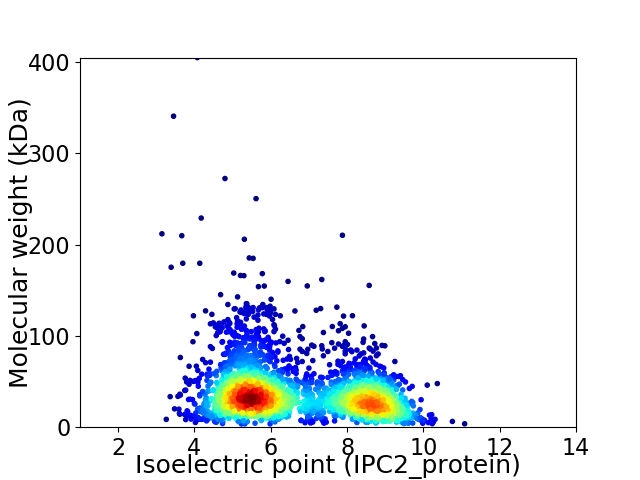
Virtual 2D-PAGE plot for 2965 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3D9H939|A0A3D9H939_9FLAO Phytoene desaturase OS=Seonamhaeicola aphaedonensis OX=1461338 GN=DFQ02_107137 PE=3 SV=1
MM1 pKa = 7.38KK2 pKa = 10.27LEE4 pKa = 4.57KK5 pKa = 10.06IGLFALCMVFCFLSCNKK22 pKa = 9.71NDD24 pKa = 3.74EE25 pKa = 4.5ASDD28 pKa = 3.65DD29 pKa = 4.24VIVVEE34 pKa = 4.37IRR36 pKa = 11.84DD37 pKa = 3.65RR38 pKa = 11.84YY39 pKa = 7.95EE40 pKa = 3.43QQIIDD45 pKa = 3.8NDD47 pKa = 4.05SLVEE51 pKa = 3.96FLEE54 pKa = 3.83THH56 pKa = 6.64YY57 pKa = 10.88YY58 pKa = 9.62NKK60 pKa = 10.48SAFEE64 pKa = 4.39GNSNPKK70 pKa = 8.53ITDD73 pKa = 3.53LQILSTQGLSISEE86 pKa = 4.26NADD89 pKa = 3.22SLLINAVGTSKK100 pKa = 9.53KK101 pKa = 8.93TVFADD106 pKa = 3.36ANYY109 pKa = 10.5EE110 pKa = 4.08YY111 pKa = 11.3YY112 pKa = 10.02ILNLNNGGGAEE123 pKa = 4.32SPTFADD129 pKa = 3.94NVLATYY135 pKa = 9.97EE136 pKa = 4.83GYY138 pKa = 8.38TLDD141 pKa = 5.1ADD143 pKa = 5.33LNNLDD148 pKa = 4.74DD149 pKa = 5.48AFDD152 pKa = 4.01SKK154 pKa = 11.66VNPDD158 pKa = 3.32TYY160 pKa = 11.45FDD162 pKa = 4.25LTTLIPGWRR171 pKa = 11.84RR172 pKa = 11.84VFPQFNVAAGYY183 pKa = 10.27VDD185 pKa = 4.7NGDD188 pKa = 3.63GTISFSNPGIGVMFLPSGLGYY209 pKa = 9.85YY210 pKa = 10.42QNAQSLIPAYY220 pKa = 10.57SPLIFKK226 pKa = 10.67FEE228 pKa = 5.06LIATTEE234 pKa = 3.96NDD236 pKa = 3.59HH237 pKa = 7.39DD238 pKa = 4.23NDD240 pKa = 5.23GIPSHH245 pKa = 7.4KK246 pKa = 10.21EE247 pKa = 3.6DD248 pKa = 5.17LNDD251 pKa = 5.52DD252 pKa = 3.52GDD254 pKa = 4.33FNINSAIGVIDD265 pKa = 4.9GDD267 pKa = 4.13DD268 pKa = 3.65TDD270 pKa = 4.67EE271 pKa = 6.0DD272 pKa = 4.88GIPNYY277 pKa = 10.04FDD279 pKa = 5.62PDD281 pKa = 4.0DD282 pKa = 5.68DD283 pKa = 5.88GDD285 pKa = 4.8GIPTANEE292 pKa = 4.84DD293 pKa = 3.25IDD295 pKa = 5.58GDD297 pKa = 4.05GDD299 pKa = 3.69PTNDD303 pKa = 2.9IGKK306 pKa = 9.68NGIAKK311 pKa = 10.23YY312 pKa = 10.29LDD314 pKa = 3.81PEE316 pKa = 4.35EE317 pKa = 4.67TEE319 pKa = 4.58SNN321 pKa = 3.47
MM1 pKa = 7.38KK2 pKa = 10.27LEE4 pKa = 4.57KK5 pKa = 10.06IGLFALCMVFCFLSCNKK22 pKa = 9.71NDD24 pKa = 3.74EE25 pKa = 4.5ASDD28 pKa = 3.65DD29 pKa = 4.24VIVVEE34 pKa = 4.37IRR36 pKa = 11.84DD37 pKa = 3.65RR38 pKa = 11.84YY39 pKa = 7.95EE40 pKa = 3.43QQIIDD45 pKa = 3.8NDD47 pKa = 4.05SLVEE51 pKa = 3.96FLEE54 pKa = 3.83THH56 pKa = 6.64YY57 pKa = 10.88YY58 pKa = 9.62NKK60 pKa = 10.48SAFEE64 pKa = 4.39GNSNPKK70 pKa = 8.53ITDD73 pKa = 3.53LQILSTQGLSISEE86 pKa = 4.26NADD89 pKa = 3.22SLLINAVGTSKK100 pKa = 9.53KK101 pKa = 8.93TVFADD106 pKa = 3.36ANYY109 pKa = 10.5EE110 pKa = 4.08YY111 pKa = 11.3YY112 pKa = 10.02ILNLNNGGGAEE123 pKa = 4.32SPTFADD129 pKa = 3.94NVLATYY135 pKa = 9.97EE136 pKa = 4.83GYY138 pKa = 8.38TLDD141 pKa = 5.1ADD143 pKa = 5.33LNNLDD148 pKa = 4.74DD149 pKa = 5.48AFDD152 pKa = 4.01SKK154 pKa = 11.66VNPDD158 pKa = 3.32TYY160 pKa = 11.45FDD162 pKa = 4.25LTTLIPGWRR171 pKa = 11.84RR172 pKa = 11.84VFPQFNVAAGYY183 pKa = 10.27VDD185 pKa = 4.7NGDD188 pKa = 3.63GTISFSNPGIGVMFLPSGLGYY209 pKa = 9.85YY210 pKa = 10.42QNAQSLIPAYY220 pKa = 10.57SPLIFKK226 pKa = 10.67FEE228 pKa = 5.06LIATTEE234 pKa = 3.96NDD236 pKa = 3.59HH237 pKa = 7.39DD238 pKa = 4.23NDD240 pKa = 5.23GIPSHH245 pKa = 7.4KK246 pKa = 10.21EE247 pKa = 3.6DD248 pKa = 5.17LNDD251 pKa = 5.52DD252 pKa = 3.52GDD254 pKa = 4.33FNINSAIGVIDD265 pKa = 4.9GDD267 pKa = 4.13DD268 pKa = 3.65TDD270 pKa = 4.67EE271 pKa = 6.0DD272 pKa = 4.88GIPNYY277 pKa = 10.04FDD279 pKa = 5.62PDD281 pKa = 4.0DD282 pKa = 5.68DD283 pKa = 5.88GDD285 pKa = 4.8GIPTANEE292 pKa = 4.84DD293 pKa = 3.25IDD295 pKa = 5.58GDD297 pKa = 4.05GDD299 pKa = 3.69PTNDD303 pKa = 2.9IGKK306 pKa = 9.68NGIAKK311 pKa = 10.23YY312 pKa = 10.29LDD314 pKa = 3.81PEE316 pKa = 4.35EE317 pKa = 4.67TEE319 pKa = 4.58SNN321 pKa = 3.47
Molecular weight: 35.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3D9H1K3|A0A3D9H1K3_9FLAO Uncharacterized protein OS=Seonamhaeicola aphaedonensis OX=1461338 GN=DFQ02_1382 PE=4 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.21KK16 pKa = 10.3RR17 pKa = 11.84KK18 pKa = 9.62KK19 pKa = 9.04RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.77KK27 pKa = 10.58KK28 pKa = 9.8KK29 pKa = 10.51KK30 pKa = 10.04
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.21KK16 pKa = 10.3RR17 pKa = 11.84KK18 pKa = 9.62KK19 pKa = 9.04RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.77KK27 pKa = 10.58KK28 pKa = 9.8KK29 pKa = 10.51KK30 pKa = 10.04
Molecular weight: 3.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1052557 |
30 |
3606 |
355.0 |
40.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.03 ± 0.044 | 0.793 ± 0.015 |
5.67 ± 0.039 | 6.535 ± 0.046 |
5.283 ± 0.038 | 6.356 ± 0.04 |
1.858 ± 0.023 | 8.034 ± 0.045 |
7.988 ± 0.063 | 9.184 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.153 ± 0.023 | 6.448 ± 0.053 |
3.418 ± 0.026 | 3.229 ± 0.025 |
3.407 ± 0.032 | 6.459 ± 0.043 |
5.647 ± 0.042 | 6.199 ± 0.035 |
1.132 ± 0.022 | 4.177 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
