
Flavobacteriales bacterium ALC-1
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; unclassified Flavobacteriales
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
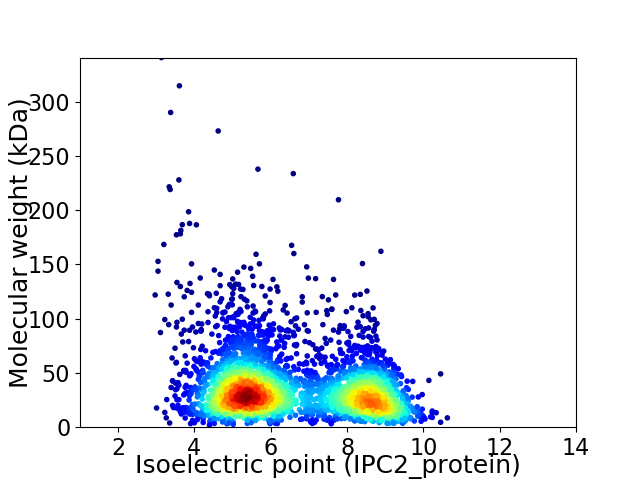
Virtual 2D-PAGE plot for 3427 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8UHX2|A8UHX2_9FLAO Putative transporter permease protein OS=Flavobacteriales bacterium ALC-1 OX=391603 GN=FBALC1_04537 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 10.39RR3 pKa = 11.84IVYY6 pKa = 10.23LLLLLPFTISAQFSVSSTTPASNEE30 pKa = 3.58LDD32 pKa = 3.47VSVTSDD38 pKa = 2.47ISINFNQNVDD48 pKa = 3.34AATLTNANISITGNNGIVLGFSAPIVTGTNAIFNPDD84 pKa = 3.03RR85 pKa = 11.84DD86 pKa = 4.11FFPGEE91 pKa = 4.39IITITITTDD100 pKa = 3.02VEE102 pKa = 4.32NSSGTNLNKK111 pKa = 10.44AFTFQFTAEE120 pKa = 4.1VCLNSPGQFPLGEE133 pKa = 4.2NTISLATDD141 pKa = 3.93GAISVTIADD150 pKa = 4.84LDD152 pKa = 4.19DD153 pKa = 6.36DD154 pKa = 5.09GDD156 pKa = 4.55LDD158 pKa = 3.83ILSASSLDD166 pKa = 5.78DD167 pKa = 4.5RR168 pKa = 11.84IAWYY172 pKa = 10.68ANDD175 pKa = 3.79GTGKK179 pKa = 10.5FGVQQTITTSADD191 pKa = 3.07YY192 pKa = 10.97AHH194 pKa = 7.52RR195 pKa = 11.84VTTADD200 pKa = 3.93LDD202 pKa = 4.02GDD204 pKa = 3.95GDD206 pKa = 3.86IDD208 pKa = 4.28VISASSVDD216 pKa = 5.72DD217 pKa = 4.58RR218 pKa = 11.84IAWYY222 pKa = 10.31EE223 pKa = 3.61NDD225 pKa = 3.27GLGNFGPQQTITTAANLAYY244 pKa = 10.49NVIASDD250 pKa = 4.45LDD252 pKa = 3.28QDD254 pKa = 3.42GDD256 pKa = 3.63MDD258 pKa = 4.78VISASAFDD266 pKa = 5.68GLIAWYY272 pKa = 10.15EE273 pKa = 3.76NDD275 pKa = 3.21GLGNFGALQTITTAAPGARR294 pKa = 11.84SVYY297 pKa = 10.3IADD300 pKa = 4.69LDD302 pKa = 3.85NDD304 pKa = 3.66GDD306 pKa = 4.07IDD308 pKa = 4.13VLSASSDD315 pKa = 3.56DD316 pKa = 5.91DD317 pKa = 3.51RR318 pKa = 11.84VAWYY322 pKa = 10.17EE323 pKa = 3.71NDD325 pKa = 3.16GSGNFGAQQTITTLADD341 pKa = 3.21RR342 pKa = 11.84AFSVKK347 pKa = 10.41AADD350 pKa = 4.12LDD352 pKa = 4.11GDD354 pKa = 3.7GDD356 pKa = 3.9MDD358 pKa = 4.81VIAACSSTSTARR370 pKa = 11.84DD371 pKa = 3.23WRR373 pKa = 11.84ITWYY377 pKa = 10.23EE378 pKa = 3.82NLGSGTFGPQQTISINVFQARR399 pKa = 11.84SVYY402 pKa = 9.77TADD405 pKa = 4.59LDD407 pKa = 4.21GDD409 pKa = 3.87GDD411 pKa = 4.97LDD413 pKa = 3.86VLSASSRR420 pKa = 11.84DD421 pKa = 4.32DD422 pKa = 4.02RR423 pKa = 11.84IAWYY427 pKa = 9.79EE428 pKa = 3.75NDD430 pKa = 3.31GSGSFGAQQTVTTNADD446 pKa = 3.7GAWSIVAADD455 pKa = 4.4LDD457 pKa = 4.31GDD459 pKa = 3.94GDD461 pKa = 3.83MDD463 pKa = 6.35LISGDD468 pKa = 3.75YY469 pKa = 11.02NGDD472 pKa = 3.09RR473 pKa = 11.84VAWYY477 pKa = 9.54EE478 pKa = 4.0NAVHH482 pKa = 7.77DD483 pKa = 4.98DD484 pKa = 3.55RR485 pKa = 11.84TLATLGGTDD494 pKa = 4.56LVLSPPFDD502 pKa = 4.92PNVSSYY508 pKa = 9.53TAAVCNSVLSTTINATVTNSGDD530 pKa = 3.87TIGGTGLVNLNEE542 pKa = 4.5GTNMITVSVTDD553 pKa = 3.39QCGGTDD559 pKa = 3.65DD560 pKa = 3.94YY561 pKa = 10.87TVEE564 pKa = 4.16VTRR567 pKa = 11.84VPLITQQITWSGSVSTDD584 pKa = 2.42WNNPNNWTPNIVPLEE599 pKa = 4.1CLEE602 pKa = 4.37IIIPEE607 pKa = 4.32TTNQPLISPGVINIKK622 pKa = 10.2DD623 pKa = 3.66LNINSGSNLEE633 pKa = 4.25IPVGATLNVSDD644 pKa = 5.05DD645 pKa = 3.71LNMYY649 pKa = 10.15SEE651 pKa = 4.78SDD653 pKa = 3.64TYY655 pKa = 11.52SGIIVKK661 pKa = 10.24GVLSVSGKK669 pKa = 8.02TKK671 pKa = 7.73YY672 pKa = 9.97HH673 pKa = 7.36RR674 pKa = 11.84YY675 pKa = 8.04TNSQANGNDD684 pKa = 4.31LIAPPLSGQSWTSFLTNDD702 pKa = 3.27ANHH705 pKa = 6.52NEE707 pKa = 3.61GLIYY711 pKa = 10.84NNGANPVTTYY721 pKa = 11.29LFGPFEE727 pKa = 4.72KK728 pKa = 10.74GATDD732 pKa = 5.79DD733 pKa = 4.27YY734 pKa = 11.6ILYY737 pKa = 10.02NDD739 pKa = 4.09NSSEE743 pKa = 3.9FLISGKK749 pKa = 9.67GYY751 pKa = 10.22RR752 pKa = 11.84SATNTVNGDD761 pKa = 3.45ALIFTGSIVTGAVNTTIINEE781 pKa = 4.36TTGGFPEE788 pKa = 4.43WNLIGNPYY796 pKa = 8.23PTYY799 pKa = 10.44IDD801 pKa = 3.46VNTFLNHH808 pKa = 5.82VGSVSGVTNLSLLSDD823 pKa = 3.84STAAVYY829 pKa = 9.92GYY831 pKa = 10.69DD832 pKa = 4.41GDD834 pKa = 4.14DD835 pKa = 3.57TDD837 pKa = 4.16VSGSVWTYY845 pKa = 8.32TNLVEE850 pKa = 4.82GPTLIAPGQGFFVSSKK866 pKa = 9.05YY867 pKa = 10.58ASSNIEE873 pKa = 4.05FTPDD877 pKa = 3.19MQIKK881 pKa = 10.24GSSDD885 pKa = 3.68DD886 pKa = 4.18FIQGRR891 pKa = 11.84TINNTDD897 pKa = 3.87YY898 pKa = 11.17IKK900 pKa = 11.21LNAATSSKK908 pKa = 10.71SYY910 pKa = 8.74STSFYY915 pKa = 10.18FHH917 pKa = 7.08NNGSQGLDD925 pKa = 2.99KK926 pKa = 11.17GYY928 pKa = 10.66DD929 pKa = 3.16AAVFGGALPSFSLYY943 pKa = 10.49SHH945 pKa = 6.99LVAEE949 pKa = 4.6NEE951 pKa = 4.42GIPMVIQTLSSDD963 pKa = 4.0DD964 pKa = 3.57IGNVIVPLGLNVNNGEE980 pKa = 4.1QVTISLGNYY989 pKa = 7.69NIPDD993 pKa = 3.59NVSVYY998 pKa = 11.14LEE1000 pKa = 4.65DD1001 pKa = 3.72NVNNTFTRR1009 pKa = 11.84LDD1011 pKa = 3.39TTDD1014 pKa = 3.95YY1015 pKa = 11.26IFTTVTALNGTGRR1028 pKa = 11.84FYY1030 pKa = 11.54LHH1032 pKa = 6.64FTTEE1036 pKa = 4.36SLLSTGEE1043 pKa = 4.15HH1044 pKa = 5.93NFKK1047 pKa = 10.62EE1048 pKa = 4.18FNIYY1052 pKa = 9.44TNQLGRR1058 pKa = 11.84TIIIEE1063 pKa = 4.17GNLTNKK1069 pKa = 9.6VDD1071 pKa = 3.72VVVFDD1076 pKa = 3.35ILGRR1080 pKa = 11.84EE1081 pKa = 4.6VLRR1084 pKa = 11.84QDD1086 pKa = 5.03LNHH1089 pKa = 6.78LVNKK1093 pKa = 10.36NIINANTLGSGVYY1106 pKa = 10.36VITIEE1111 pKa = 4.75DD1112 pKa = 3.67GNRR1115 pKa = 11.84QYY1117 pKa = 11.0SQKK1120 pKa = 10.85LILKK1124 pKa = 9.44
MM1 pKa = 7.28KK2 pKa = 10.39RR3 pKa = 11.84IVYY6 pKa = 10.23LLLLLPFTISAQFSVSSTTPASNEE30 pKa = 3.58LDD32 pKa = 3.47VSVTSDD38 pKa = 2.47ISINFNQNVDD48 pKa = 3.34AATLTNANISITGNNGIVLGFSAPIVTGTNAIFNPDD84 pKa = 3.03RR85 pKa = 11.84DD86 pKa = 4.11FFPGEE91 pKa = 4.39IITITITTDD100 pKa = 3.02VEE102 pKa = 4.32NSSGTNLNKK111 pKa = 10.44AFTFQFTAEE120 pKa = 4.1VCLNSPGQFPLGEE133 pKa = 4.2NTISLATDD141 pKa = 3.93GAISVTIADD150 pKa = 4.84LDD152 pKa = 4.19DD153 pKa = 6.36DD154 pKa = 5.09GDD156 pKa = 4.55LDD158 pKa = 3.83ILSASSLDD166 pKa = 5.78DD167 pKa = 4.5RR168 pKa = 11.84IAWYY172 pKa = 10.68ANDD175 pKa = 3.79GTGKK179 pKa = 10.5FGVQQTITTSADD191 pKa = 3.07YY192 pKa = 10.97AHH194 pKa = 7.52RR195 pKa = 11.84VTTADD200 pKa = 3.93LDD202 pKa = 4.02GDD204 pKa = 3.95GDD206 pKa = 3.86IDD208 pKa = 4.28VISASSVDD216 pKa = 5.72DD217 pKa = 4.58RR218 pKa = 11.84IAWYY222 pKa = 10.31EE223 pKa = 3.61NDD225 pKa = 3.27GLGNFGPQQTITTAANLAYY244 pKa = 10.49NVIASDD250 pKa = 4.45LDD252 pKa = 3.28QDD254 pKa = 3.42GDD256 pKa = 3.63MDD258 pKa = 4.78VISASAFDD266 pKa = 5.68GLIAWYY272 pKa = 10.15EE273 pKa = 3.76NDD275 pKa = 3.21GLGNFGALQTITTAAPGARR294 pKa = 11.84SVYY297 pKa = 10.3IADD300 pKa = 4.69LDD302 pKa = 3.85NDD304 pKa = 3.66GDD306 pKa = 4.07IDD308 pKa = 4.13VLSASSDD315 pKa = 3.56DD316 pKa = 5.91DD317 pKa = 3.51RR318 pKa = 11.84VAWYY322 pKa = 10.17EE323 pKa = 3.71NDD325 pKa = 3.16GSGNFGAQQTITTLADD341 pKa = 3.21RR342 pKa = 11.84AFSVKK347 pKa = 10.41AADD350 pKa = 4.12LDD352 pKa = 4.11GDD354 pKa = 3.7GDD356 pKa = 3.9MDD358 pKa = 4.81VIAACSSTSTARR370 pKa = 11.84DD371 pKa = 3.23WRR373 pKa = 11.84ITWYY377 pKa = 10.23EE378 pKa = 3.82NLGSGTFGPQQTISINVFQARR399 pKa = 11.84SVYY402 pKa = 9.77TADD405 pKa = 4.59LDD407 pKa = 4.21GDD409 pKa = 3.87GDD411 pKa = 4.97LDD413 pKa = 3.86VLSASSRR420 pKa = 11.84DD421 pKa = 4.32DD422 pKa = 4.02RR423 pKa = 11.84IAWYY427 pKa = 9.79EE428 pKa = 3.75NDD430 pKa = 3.31GSGSFGAQQTVTTNADD446 pKa = 3.7GAWSIVAADD455 pKa = 4.4LDD457 pKa = 4.31GDD459 pKa = 3.94GDD461 pKa = 3.83MDD463 pKa = 6.35LISGDD468 pKa = 3.75YY469 pKa = 11.02NGDD472 pKa = 3.09RR473 pKa = 11.84VAWYY477 pKa = 9.54EE478 pKa = 4.0NAVHH482 pKa = 7.77DD483 pKa = 4.98DD484 pKa = 3.55RR485 pKa = 11.84TLATLGGTDD494 pKa = 4.56LVLSPPFDD502 pKa = 4.92PNVSSYY508 pKa = 9.53TAAVCNSVLSTTINATVTNSGDD530 pKa = 3.87TIGGTGLVNLNEE542 pKa = 4.5GTNMITVSVTDD553 pKa = 3.39QCGGTDD559 pKa = 3.65DD560 pKa = 3.94YY561 pKa = 10.87TVEE564 pKa = 4.16VTRR567 pKa = 11.84VPLITQQITWSGSVSTDD584 pKa = 2.42WNNPNNWTPNIVPLEE599 pKa = 4.1CLEE602 pKa = 4.37IIIPEE607 pKa = 4.32TTNQPLISPGVINIKK622 pKa = 10.2DD623 pKa = 3.66LNINSGSNLEE633 pKa = 4.25IPVGATLNVSDD644 pKa = 5.05DD645 pKa = 3.71LNMYY649 pKa = 10.15SEE651 pKa = 4.78SDD653 pKa = 3.64TYY655 pKa = 11.52SGIIVKK661 pKa = 10.24GVLSVSGKK669 pKa = 8.02TKK671 pKa = 7.73YY672 pKa = 9.97HH673 pKa = 7.36RR674 pKa = 11.84YY675 pKa = 8.04TNSQANGNDD684 pKa = 4.31LIAPPLSGQSWTSFLTNDD702 pKa = 3.27ANHH705 pKa = 6.52NEE707 pKa = 3.61GLIYY711 pKa = 10.84NNGANPVTTYY721 pKa = 11.29LFGPFEE727 pKa = 4.72KK728 pKa = 10.74GATDD732 pKa = 5.79DD733 pKa = 4.27YY734 pKa = 11.6ILYY737 pKa = 10.02NDD739 pKa = 4.09NSSEE743 pKa = 3.9FLISGKK749 pKa = 9.67GYY751 pKa = 10.22RR752 pKa = 11.84SATNTVNGDD761 pKa = 3.45ALIFTGSIVTGAVNTTIINEE781 pKa = 4.36TTGGFPEE788 pKa = 4.43WNLIGNPYY796 pKa = 8.23PTYY799 pKa = 10.44IDD801 pKa = 3.46VNTFLNHH808 pKa = 5.82VGSVSGVTNLSLLSDD823 pKa = 3.84STAAVYY829 pKa = 9.92GYY831 pKa = 10.69DD832 pKa = 4.41GDD834 pKa = 4.14DD835 pKa = 3.57TDD837 pKa = 4.16VSGSVWTYY845 pKa = 8.32TNLVEE850 pKa = 4.82GPTLIAPGQGFFVSSKK866 pKa = 9.05YY867 pKa = 10.58ASSNIEE873 pKa = 4.05FTPDD877 pKa = 3.19MQIKK881 pKa = 10.24GSSDD885 pKa = 3.68DD886 pKa = 4.18FIQGRR891 pKa = 11.84TINNTDD897 pKa = 3.87YY898 pKa = 11.17IKK900 pKa = 11.21LNAATSSKK908 pKa = 10.71SYY910 pKa = 8.74STSFYY915 pKa = 10.18FHH917 pKa = 7.08NNGSQGLDD925 pKa = 2.99KK926 pKa = 11.17GYY928 pKa = 10.66DD929 pKa = 3.16AAVFGGALPSFSLYY943 pKa = 10.49SHH945 pKa = 6.99LVAEE949 pKa = 4.6NEE951 pKa = 4.42GIPMVIQTLSSDD963 pKa = 4.0DD964 pKa = 3.57IGNVIVPLGLNVNNGEE980 pKa = 4.1QVTISLGNYY989 pKa = 7.69NIPDD993 pKa = 3.59NVSVYY998 pKa = 11.14LEE1000 pKa = 4.65DD1001 pKa = 3.72NVNNTFTRR1009 pKa = 11.84LDD1011 pKa = 3.39TTDD1014 pKa = 3.95YY1015 pKa = 11.26IFTTVTALNGTGRR1028 pKa = 11.84FYY1030 pKa = 11.54LHH1032 pKa = 6.64FTTEE1036 pKa = 4.36SLLSTGEE1043 pKa = 4.15HH1044 pKa = 5.93NFKK1047 pKa = 10.62EE1048 pKa = 4.18FNIYY1052 pKa = 9.44TNQLGRR1058 pKa = 11.84TIIIEE1063 pKa = 4.17GNLTNKK1069 pKa = 9.6VDD1071 pKa = 3.72VVVFDD1076 pKa = 3.35ILGRR1080 pKa = 11.84EE1081 pKa = 4.6VLRR1084 pKa = 11.84QDD1086 pKa = 5.03LNHH1089 pKa = 6.78LVNKK1093 pKa = 10.36NIINANTLGSGVYY1106 pKa = 10.36VITIEE1111 pKa = 4.75DD1112 pKa = 3.67GNRR1115 pKa = 11.84QYY1117 pKa = 11.0SQKK1120 pKa = 10.85LILKK1124 pKa = 9.44
Molecular weight: 120.23 kDa
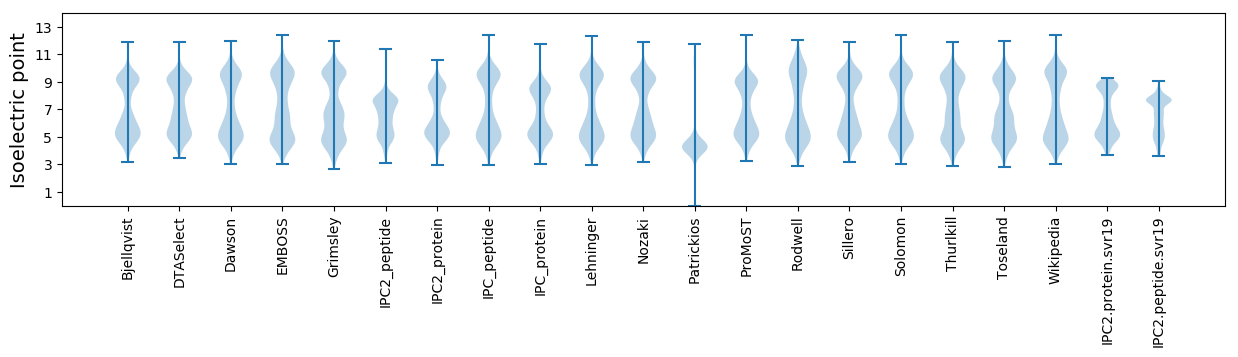
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8UH16|A8UH16_9FLAO Transcriptional repressor of sporulation and degradative enzyme production OS=Flavobacteriales bacterium ALC-1 OX=391603 GN=FBALC1_13512 PE=4 SV=1
MM1 pKa = 7.3ARR3 pKa = 11.84IAGVDD8 pKa = 2.98IPKK11 pKa = 10.23NKK13 pKa = 9.96RR14 pKa = 11.84GVISLTYY21 pKa = 8.93IYY23 pKa = 10.94GVGRR27 pKa = 11.84SRR29 pKa = 11.84AKK31 pKa = 10.5EE32 pKa = 3.56ILATAKK38 pKa = 10.09VDD40 pKa = 3.51EE41 pKa = 4.52STKK44 pKa = 9.93VQDD47 pKa = 3.51WSDD50 pKa = 3.97DD51 pKa = 3.89EE52 pKa = 4.88IGAIRR57 pKa = 11.84EE58 pKa = 4.08AVGTFKK64 pKa = 10.66IEE66 pKa = 3.87GEE68 pKa = 4.25LRR70 pKa = 11.84SEE72 pKa = 3.86TQMNIKK78 pKa = 10.21RR79 pKa = 11.84LMDD82 pKa = 3.76IGCYY86 pKa = 9.34RR87 pKa = 11.84GIRR90 pKa = 11.84HH91 pKa = 6.13RR92 pKa = 11.84VGLPLRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 11.3NNSRR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.17GRR112 pKa = 11.84RR113 pKa = 11.84KK114 pKa = 7.81TVANKK119 pKa = 10.49KK120 pKa = 10.06KK121 pKa = 8.78VTKK124 pKa = 10.51
MM1 pKa = 7.3ARR3 pKa = 11.84IAGVDD8 pKa = 2.98IPKK11 pKa = 10.23NKK13 pKa = 9.96RR14 pKa = 11.84GVISLTYY21 pKa = 8.93IYY23 pKa = 10.94GVGRR27 pKa = 11.84SRR29 pKa = 11.84AKK31 pKa = 10.5EE32 pKa = 3.56ILATAKK38 pKa = 10.09VDD40 pKa = 3.51EE41 pKa = 4.52STKK44 pKa = 9.93VQDD47 pKa = 3.51WSDD50 pKa = 3.97DD51 pKa = 3.89EE52 pKa = 4.88IGAIRR57 pKa = 11.84EE58 pKa = 4.08AVGTFKK64 pKa = 10.66IEE66 pKa = 3.87GEE68 pKa = 4.25LRR70 pKa = 11.84SEE72 pKa = 3.86TQMNIKK78 pKa = 10.21RR79 pKa = 11.84LMDD82 pKa = 3.76IGCYY86 pKa = 9.34RR87 pKa = 11.84GIRR90 pKa = 11.84HH91 pKa = 6.13RR92 pKa = 11.84VGLPLRR98 pKa = 11.84GQRR101 pKa = 11.84TKK103 pKa = 11.3NNSRR107 pKa = 11.84TRR109 pKa = 11.84KK110 pKa = 9.17GRR112 pKa = 11.84RR113 pKa = 11.84KK114 pKa = 7.81TVANKK119 pKa = 10.49KK120 pKa = 10.06KK121 pKa = 8.78VTKK124 pKa = 10.51
Molecular weight: 14.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1172252 |
17 |
3247 |
342.1 |
38.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.138 ± 0.036 | 0.745 ± 0.017 |
5.977 ± 0.039 | 6.374 ± 0.038 |
5.215 ± 0.036 | 6.266 ± 0.053 |
1.694 ± 0.019 | 8.239 ± 0.043 |
7.817 ± 0.067 | 9.179 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.093 ± 0.02 | 6.612 ± 0.047 |
3.216 ± 0.027 | 3.185 ± 0.022 |
3.248 ± 0.029 | 6.748 ± 0.034 |
5.963 ± 0.059 | 6.1 ± 0.028 |
1.033 ± 0.016 | 4.16 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
