
Novosphingobium sp. Fuku2-ISO-50
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium; unclassified Novosphingobium
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
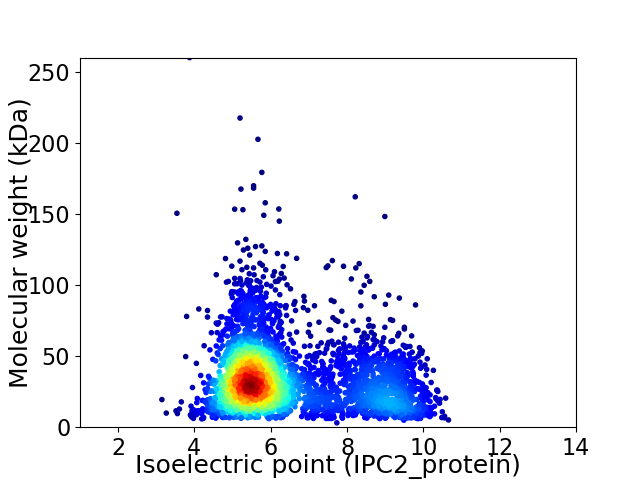
Virtual 2D-PAGE plot for 3834 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A117V1C9|A0A117V1C9_9SPHN Uncharacterized protein OS=Novosphingobium sp. Fuku2-ISO-50 OX=1739114 GN=AQZ50_16665 PE=3 SV=1
MM1 pKa = 7.7SYY3 pKa = 10.78AISLSGLQNAQTDD16 pKa = 3.84LSVIGNNIANANTTGFKK33 pKa = 10.24EE34 pKa = 4.67SNVQFANLVAASAYY48 pKa = 10.67SNPKK52 pKa = 9.53DD53 pKa = 3.31IQGIGSTVSAIDD65 pKa = 3.36QNFSQGPINQTGSALDD81 pKa = 3.7LAITGDD87 pKa = 3.72GFFTVTSPITTQTYY101 pKa = 6.14YY102 pKa = 10.46TRR104 pKa = 11.84NGNFSLDD111 pKa = 3.41STTAATSGNSYY122 pKa = 10.51IVDD125 pKa = 3.73SNGNRR130 pKa = 11.84LQALPTSATGTVTSTTPTDD149 pKa = 3.41ATVPATDD156 pKa = 3.47AAGSAFSGVTIAANGNISASYY177 pKa = 10.84ADD179 pKa = 3.91GSNTVIGKK187 pKa = 9.08VALAAFAAPEE197 pKa = 4.11GLKK200 pKa = 10.28QVGSADD206 pKa = 3.53YY207 pKa = 8.63TATGLSGTATFGAPNSGQYY226 pKa = 11.09GSLLSGSLEE235 pKa = 3.87ASNVDD240 pKa = 3.84LSSEE244 pKa = 4.15MVNLITAQQYY254 pKa = 8.12FQANAKK260 pKa = 10.49AIDD263 pKa = 3.75TNTTVSEE270 pKa = 4.52AIINLHH276 pKa = 5.47SS277 pKa = 3.61
MM1 pKa = 7.7SYY3 pKa = 10.78AISLSGLQNAQTDD16 pKa = 3.84LSVIGNNIANANTTGFKK33 pKa = 10.24EE34 pKa = 4.67SNVQFANLVAASAYY48 pKa = 10.67SNPKK52 pKa = 9.53DD53 pKa = 3.31IQGIGSTVSAIDD65 pKa = 3.36QNFSQGPINQTGSALDD81 pKa = 3.7LAITGDD87 pKa = 3.72GFFTVTSPITTQTYY101 pKa = 6.14YY102 pKa = 10.46TRR104 pKa = 11.84NGNFSLDD111 pKa = 3.41STTAATSGNSYY122 pKa = 10.51IVDD125 pKa = 3.73SNGNRR130 pKa = 11.84LQALPTSATGTVTSTTPTDD149 pKa = 3.41ATVPATDD156 pKa = 3.47AAGSAFSGVTIAANGNISASYY177 pKa = 10.84ADD179 pKa = 3.91GSNTVIGKK187 pKa = 9.08VALAAFAAPEE197 pKa = 4.11GLKK200 pKa = 10.28QVGSADD206 pKa = 3.53YY207 pKa = 8.63TATGLSGTATFGAPNSGQYY226 pKa = 11.09GSLLSGSLEE235 pKa = 3.87ASNVDD240 pKa = 3.84LSSEE244 pKa = 4.15MVNLITAQQYY254 pKa = 8.12FQANAKK260 pKa = 10.49AIDD263 pKa = 3.75TNTTVSEE270 pKa = 4.52AIINLHH276 pKa = 5.47SS277 pKa = 3.61
Molecular weight: 27.95 kDa
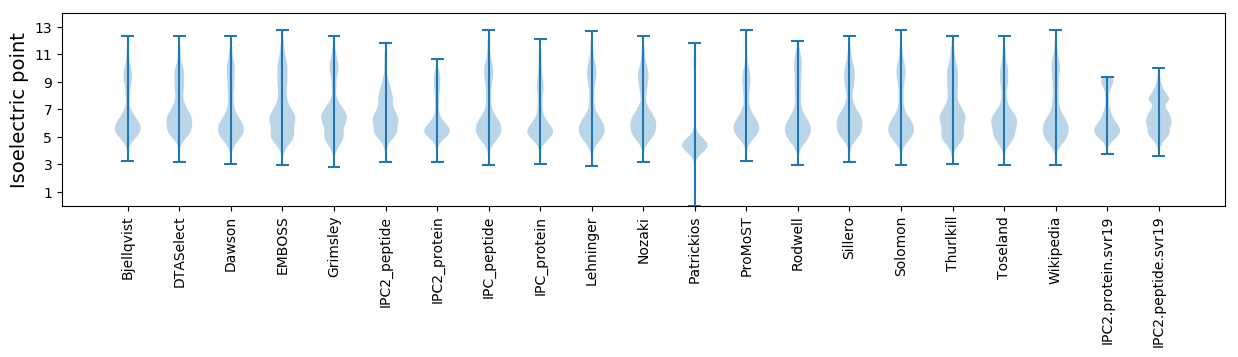
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A102DF85|A0A102DF85_9SPHN Dihydroorotase OS=Novosphingobium sp. Fuku2-ISO-50 OX=1739114 GN=pyrC PE=3 SV=1
MM1 pKa = 7.81RR2 pKa = 11.84IEE4 pKa = 4.08RR5 pKa = 11.84MNGRR9 pKa = 11.84GLRR12 pKa = 11.84MTSATPLRR20 pKa = 11.84VVAASALLLAAGGCSTIRR38 pKa = 11.84DD39 pKa = 3.58HH40 pKa = 7.74RR41 pKa = 11.84GYY43 pKa = 10.89LVDD46 pKa = 3.93PALTQSVLPGVDD58 pKa = 2.85NRR60 pKa = 11.84LSVEE64 pKa = 4.01KK65 pKa = 10.92SLGQPTLKK73 pKa = 10.66SQFGQQSWYY82 pKa = 10.0YY83 pKa = 10.19VSLDD87 pKa = 3.29TVQRR91 pKa = 11.84PFTRR95 pKa = 11.84PRR97 pKa = 11.84IHH99 pKa = 6.83AGSILRR105 pKa = 11.84VSFDD109 pKa = 3.15TAGTVQSVSNDD120 pKa = 3.31GVEE123 pKa = 3.88HH124 pKa = 6.6AQRR127 pKa = 11.84IVPDD131 pKa = 3.38RR132 pKa = 11.84HH133 pKa = 5.21KK134 pKa = 9.82TPTLGRR140 pKa = 11.84DD141 pKa = 2.74RR142 pKa = 11.84SFFEE146 pKa = 5.36DD147 pKa = 3.31LFGNIGTVGAMPGAQGGQPGGGGGGRR173 pKa = 11.84GGGGTGPNGSS183 pKa = 3.72
MM1 pKa = 7.81RR2 pKa = 11.84IEE4 pKa = 4.08RR5 pKa = 11.84MNGRR9 pKa = 11.84GLRR12 pKa = 11.84MTSATPLRR20 pKa = 11.84VVAASALLLAAGGCSTIRR38 pKa = 11.84DD39 pKa = 3.58HH40 pKa = 7.74RR41 pKa = 11.84GYY43 pKa = 10.89LVDD46 pKa = 3.93PALTQSVLPGVDD58 pKa = 2.85NRR60 pKa = 11.84LSVEE64 pKa = 4.01KK65 pKa = 10.92SLGQPTLKK73 pKa = 10.66SQFGQQSWYY82 pKa = 10.0YY83 pKa = 10.19VSLDD87 pKa = 3.29TVQRR91 pKa = 11.84PFTRR95 pKa = 11.84PRR97 pKa = 11.84IHH99 pKa = 6.83AGSILRR105 pKa = 11.84VSFDD109 pKa = 3.15TAGTVQSVSNDD120 pKa = 3.31GVEE123 pKa = 3.88HH124 pKa = 6.6AQRR127 pKa = 11.84IVPDD131 pKa = 3.38RR132 pKa = 11.84HH133 pKa = 5.21KK134 pKa = 9.82TPTLGRR140 pKa = 11.84DD141 pKa = 2.74RR142 pKa = 11.84SFFEE146 pKa = 5.36DD147 pKa = 3.31LFGNIGTVGAMPGAQGGQPGGGGGGRR173 pKa = 11.84GGGGTGPNGSS183 pKa = 3.72
Molecular weight: 19.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1291571 |
29 |
2696 |
336.9 |
36.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.93 ± 0.063 | 0.853 ± 0.013 |
5.942 ± 0.029 | 4.866 ± 0.033 |
3.528 ± 0.022 | 9.091 ± 0.042 |
2.315 ± 0.018 | 4.975 ± 0.027 |
2.574 ± 0.027 | 9.911 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.407 ± 0.018 | 2.499 ± 0.029 |
5.474 ± 0.032 | 3.07 ± 0.023 |
7.174 ± 0.041 | 5.0 ± 0.033 |
5.401 ± 0.04 | 7.273 ± 0.034 |
1.481 ± 0.016 | 2.237 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
