
Bat coronavirus Cp/Yunnan2011
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Sarbecovirus; Severe acute respiratory syndrome-related coronavirus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
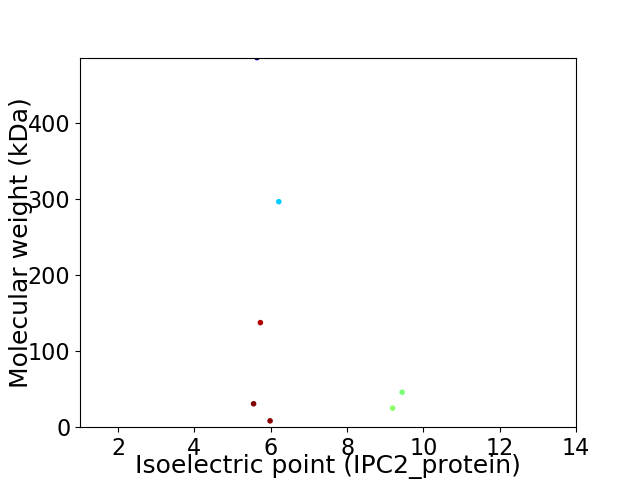
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
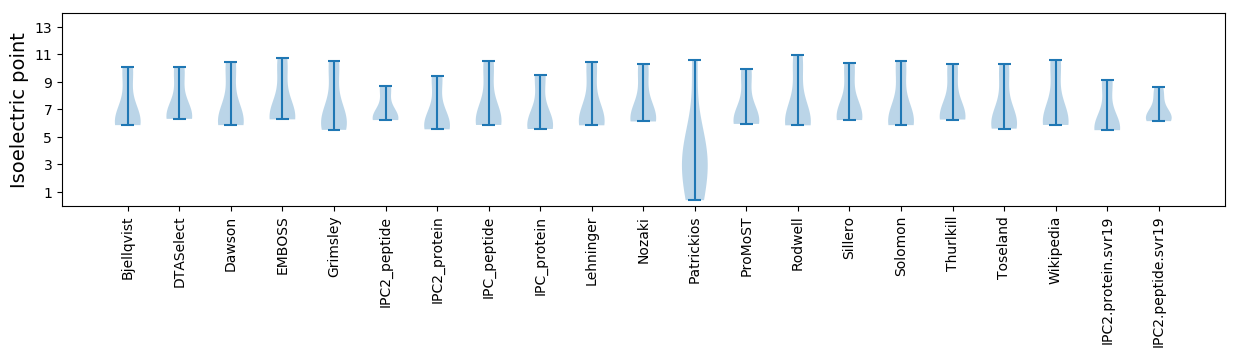
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9QTJ3|R9QTJ3_SARS ORF3 protein OS=Bat coronavirus Cp/Yunnan2011 OX=1283333 PE=4 SV=1
MM1 pKa = 7.73KK2 pKa = 10.3IFLFSLLFSAALAQEE17 pKa = 4.63GCGLLSFKK25 pKa = 10.04PQPKK29 pKa = 9.57LAQFSSSKK37 pKa = 10.5RR38 pKa = 11.84GVYY41 pKa = 10.72YY42 pKa = 10.63NDD44 pKa = 4.77DD45 pKa = 3.51IFRR48 pKa = 11.84SDD50 pKa = 3.84VLHH53 pKa = 6.33LTQDD57 pKa = 3.37YY58 pKa = 9.62FLPFHH63 pKa = 7.26SNLTQYY69 pKa = 10.92FSLNVDD75 pKa = 3.27SDD77 pKa = 3.82RR78 pKa = 11.84QVYY81 pKa = 9.95FDD83 pKa = 4.64NPTLNFGDD91 pKa = 3.83GVYY94 pKa = 10.25FAATEE99 pKa = 3.92KK100 pKa = 11.13SNVIRR105 pKa = 11.84GWIFGSTMDD114 pKa = 4.87NSTQSAIIVNNSTHH128 pKa = 7.21IIIRR132 pKa = 11.84VCNFNLCKK140 pKa = 10.5EE141 pKa = 4.14PMFTVSRR148 pKa = 11.84GVHH151 pKa = 5.37FSSWVYY157 pKa = 10.31QSAFNCTYY165 pKa = 11.31DD166 pKa = 3.42RR167 pKa = 11.84VEE169 pKa = 5.14KK170 pKa = 10.81SFQLDD175 pKa = 3.62TAPKK179 pKa = 8.49TGNFKK184 pKa = 10.73DD185 pKa = 3.24LRR187 pKa = 11.84EE188 pKa = 4.05YY189 pKa = 10.89VFKK192 pKa = 11.26NRR194 pKa = 11.84DD195 pKa = 3.16GFLSVYY201 pKa = 10.32HH202 pKa = 7.0SYY204 pKa = 10.57TPVDD208 pKa = 4.42IIRR211 pKa = 11.84GIPVGFSVLKK221 pKa = 10.32PILKK225 pKa = 10.31LPIGINITSFKK236 pKa = 11.08VVMTMYY242 pKa = 10.76SQTTSNFLSEE252 pKa = 3.95SAAYY256 pKa = 10.04YY257 pKa = 10.23VGNLKK262 pKa = 10.56YY263 pKa = 9.83VTFMFQFNEE272 pKa = 3.9NGTIADD278 pKa = 4.89AVDD281 pKa = 4.35CSQNPLAEE289 pKa = 4.46LKK291 pKa = 9.51CTLKK295 pKa = 10.83NFNVSKK301 pKa = 10.97GIYY304 pKa = 6.16QTSNFRR310 pKa = 11.84VSPSTEE316 pKa = 3.7VIRR319 pKa = 11.84FPNITNRR326 pKa = 11.84CPFDD330 pKa = 3.58RR331 pKa = 11.84VFNASRR337 pKa = 11.84FPSVYY342 pKa = 9.2AWEE345 pKa = 4.04RR346 pKa = 11.84TKK348 pKa = 10.81ISDD351 pKa = 3.69CVADD355 pKa = 3.7YY356 pKa = 9.98TVLYY360 pKa = 10.29NSTSFSTFKK369 pKa = 10.75CYY371 pKa = 10.03GVSPSKK377 pKa = 11.02LIDD380 pKa = 3.28LCFTSVYY387 pKa = 10.97ADD389 pKa = 3.22TFLIRR394 pKa = 11.84FSEE397 pKa = 4.39VRR399 pKa = 11.84QIAPGEE405 pKa = 4.15TGVIADD411 pKa = 4.42YY412 pKa = 10.83NYY414 pKa = 11.06KK415 pKa = 10.7LPDD418 pKa = 3.66EE419 pKa = 4.37FTGCVIAWNTANQDD433 pKa = 2.9RR434 pKa = 11.84GQYY437 pKa = 9.81YY438 pKa = 9.57YY439 pKa = 10.87RR440 pKa = 11.84SSRR443 pKa = 11.84KK444 pKa = 7.58TKK446 pKa = 10.3LKK448 pKa = 10.27PFEE451 pKa = 5.22RR452 pKa = 11.84DD453 pKa = 3.0LSSDD457 pKa = 3.51EE458 pKa = 4.18NGVRR462 pKa = 11.84TLSTYY467 pKa = 11.13DD468 pKa = 4.03FYY470 pKa = 11.17PSVPLEE476 pKa = 4.01YY477 pKa = 9.74QATRR481 pKa = 11.84VVVLSFEE488 pKa = 4.61LLNAPATVCGPKK500 pKa = 10.42LSTSLIKK507 pKa = 10.38NQCVNFNFNGLKK519 pKa = 9.11GTGVLTDD526 pKa = 3.51SSKK529 pKa = 11.0KK530 pKa = 9.96FQSFQQFGRR539 pKa = 11.84DD540 pKa = 3.18ASDD543 pKa = 3.34FTDD546 pKa = 3.68SVRR549 pKa = 11.84DD550 pKa = 3.75PQTLQILDD558 pKa = 3.94ISPCSFGGVSVITPGTNASSEE579 pKa = 4.47VAVLYY584 pKa = 10.73QDD586 pKa = 4.29VNCTDD591 pKa = 3.01VPTAIRR597 pKa = 11.84ADD599 pKa = 3.52QLTPAWRR606 pKa = 11.84VYY608 pKa = 10.25SAGVNVFQTQAGCLIGAEE626 pKa = 4.44HH627 pKa = 6.46VNASYY632 pKa = 11.03EE633 pKa = 4.01CDD635 pKa = 3.21IPIGAGICASYY646 pKa = 8.37HH647 pKa = 4.65TASLLRR653 pKa = 11.84NTGQKK658 pKa = 10.51SIVAYY663 pKa = 9.53TMSLGAEE670 pKa = 3.91NSIAYY675 pKa = 9.44ANNSIAIPTNFSISVTTEE693 pKa = 3.49VMPVSMAKK701 pKa = 9.66TSVDD705 pKa = 3.1CTMYY709 pKa = 10.34ICGDD713 pKa = 3.6SQEE716 pKa = 4.51CSNLLLQYY724 pKa = 11.24GSFCTQLNRR733 pKa = 11.84ALSGIAVEE741 pKa = 4.22QDD743 pKa = 2.97KK744 pKa = 8.86NTQEE748 pKa = 3.75VFAQVKK754 pKa = 9.11QMYY757 pKa = 7.06KK758 pKa = 9.37TPAIKK763 pKa = 10.61DD764 pKa = 3.33FGGFNFSQILPDD776 pKa = 3.66PSKK779 pKa = 8.47PTKK782 pKa = 10.11RR783 pKa = 11.84SFIEE787 pKa = 4.3DD788 pKa = 3.61LLFNKK793 pKa = 7.99VTLADD798 pKa = 3.57AGFMKK803 pKa = 10.37QYY805 pKa = 10.74GEE807 pKa = 4.27CLGDD811 pKa = 3.41ISARR815 pKa = 11.84DD816 pKa = 4.03LICAQKK822 pKa = 10.77FNGLTVLPPLLTDD835 pKa = 3.7EE836 pKa = 5.07MIAAYY841 pKa = 8.13TAALVSGTATAGWTFGAGAALQIPFAMQMAYY872 pKa = 10.08RR873 pKa = 11.84FNGIGVTQNVLYY885 pKa = 10.46EE886 pKa = 4.09NQKK889 pKa = 10.62QIANQFNKK897 pKa = 10.46AISQIQEE904 pKa = 4.19SLTTTSTALGKK915 pKa = 10.37LQDD918 pKa = 3.95VVNQNAQALNTLVKK932 pKa = 10.38QLSSNFGAISSVLNDD947 pKa = 3.17ILSRR951 pKa = 11.84LDD953 pKa = 3.31KK954 pKa = 11.45VEE956 pKa = 5.19AEE958 pKa = 4.28VQIDD962 pKa = 3.56RR963 pKa = 11.84LITGRR968 pKa = 11.84LQSLQTYY975 pKa = 7.4VTQQLIRR982 pKa = 11.84AAEE985 pKa = 3.85IRR987 pKa = 11.84ASANLAATKK996 pKa = 9.96MSEE999 pKa = 4.28CVLGQSKK1006 pKa = 10.12RR1007 pKa = 11.84VDD1009 pKa = 4.0FCGKK1013 pKa = 9.92GYY1015 pKa = 10.57HH1016 pKa = 6.84LMSFPQAAPHH1026 pKa = 5.76GVVFLHH1032 pKa = 5.25VTYY1035 pKa = 10.82VPSQEE1040 pKa = 4.9RR1041 pKa = 11.84NFTTAPAICHH1051 pKa = 5.3QGKK1054 pKa = 10.39AYY1056 pKa = 9.98FPRR1059 pKa = 11.84EE1060 pKa = 3.84GVFVSNGTSWFITQRR1075 pKa = 11.84NFFSPQIITTDD1086 pKa = 3.16NTFVSGNCDD1095 pKa = 3.2VVIGIINNTVYY1106 pKa = 10.97DD1107 pKa = 4.22PLQPEE1112 pKa = 4.19LDD1114 pKa = 3.85SFKK1117 pKa = 11.4EE1118 pKa = 3.91EE1119 pKa = 3.56LDD1121 pKa = 3.63KK1122 pKa = 11.53YY1123 pKa = 10.15FKK1125 pKa = 10.99NHH1127 pKa = 6.01TSPDD1131 pKa = 3.14VDD1133 pKa = 4.4LGDD1136 pKa = 3.31ISGINASVVNIQKK1149 pKa = 10.63EE1150 pKa = 3.92IDD1152 pKa = 3.67RR1153 pKa = 11.84LNEE1156 pKa = 3.5VAKK1159 pKa = 10.61NLNEE1163 pKa = 4.14SLIDD1167 pKa = 3.75LQEE1170 pKa = 4.17LGKK1173 pKa = 10.54YY1174 pKa = 6.98EE1175 pKa = 5.3QYY1177 pKa = 10.78IKK1179 pKa = 10.03WPWYY1183 pKa = 8.39VWLGFIAGLIAIVMATILLCCMTSCCSCLKK1213 pKa = 10.16GACSCGSCCKK1223 pKa = 10.16FDD1225 pKa = 4.43EE1226 pKa = 5.39DD1227 pKa = 5.38DD1228 pKa = 4.38SEE1230 pKa = 4.36PVLKK1234 pKa = 10.36GVKK1237 pKa = 9.35LHH1239 pKa = 5.3YY1240 pKa = 9.44TT1241 pKa = 3.44
MM1 pKa = 7.73KK2 pKa = 10.3IFLFSLLFSAALAQEE17 pKa = 4.63GCGLLSFKK25 pKa = 10.04PQPKK29 pKa = 9.57LAQFSSSKK37 pKa = 10.5RR38 pKa = 11.84GVYY41 pKa = 10.72YY42 pKa = 10.63NDD44 pKa = 4.77DD45 pKa = 3.51IFRR48 pKa = 11.84SDD50 pKa = 3.84VLHH53 pKa = 6.33LTQDD57 pKa = 3.37YY58 pKa = 9.62FLPFHH63 pKa = 7.26SNLTQYY69 pKa = 10.92FSLNVDD75 pKa = 3.27SDD77 pKa = 3.82RR78 pKa = 11.84QVYY81 pKa = 9.95FDD83 pKa = 4.64NPTLNFGDD91 pKa = 3.83GVYY94 pKa = 10.25FAATEE99 pKa = 3.92KK100 pKa = 11.13SNVIRR105 pKa = 11.84GWIFGSTMDD114 pKa = 4.87NSTQSAIIVNNSTHH128 pKa = 7.21IIIRR132 pKa = 11.84VCNFNLCKK140 pKa = 10.5EE141 pKa = 4.14PMFTVSRR148 pKa = 11.84GVHH151 pKa = 5.37FSSWVYY157 pKa = 10.31QSAFNCTYY165 pKa = 11.31DD166 pKa = 3.42RR167 pKa = 11.84VEE169 pKa = 5.14KK170 pKa = 10.81SFQLDD175 pKa = 3.62TAPKK179 pKa = 8.49TGNFKK184 pKa = 10.73DD185 pKa = 3.24LRR187 pKa = 11.84EE188 pKa = 4.05YY189 pKa = 10.89VFKK192 pKa = 11.26NRR194 pKa = 11.84DD195 pKa = 3.16GFLSVYY201 pKa = 10.32HH202 pKa = 7.0SYY204 pKa = 10.57TPVDD208 pKa = 4.42IIRR211 pKa = 11.84GIPVGFSVLKK221 pKa = 10.32PILKK225 pKa = 10.31LPIGINITSFKK236 pKa = 11.08VVMTMYY242 pKa = 10.76SQTTSNFLSEE252 pKa = 3.95SAAYY256 pKa = 10.04YY257 pKa = 10.23VGNLKK262 pKa = 10.56YY263 pKa = 9.83VTFMFQFNEE272 pKa = 3.9NGTIADD278 pKa = 4.89AVDD281 pKa = 4.35CSQNPLAEE289 pKa = 4.46LKK291 pKa = 9.51CTLKK295 pKa = 10.83NFNVSKK301 pKa = 10.97GIYY304 pKa = 6.16QTSNFRR310 pKa = 11.84VSPSTEE316 pKa = 3.7VIRR319 pKa = 11.84FPNITNRR326 pKa = 11.84CPFDD330 pKa = 3.58RR331 pKa = 11.84VFNASRR337 pKa = 11.84FPSVYY342 pKa = 9.2AWEE345 pKa = 4.04RR346 pKa = 11.84TKK348 pKa = 10.81ISDD351 pKa = 3.69CVADD355 pKa = 3.7YY356 pKa = 9.98TVLYY360 pKa = 10.29NSTSFSTFKK369 pKa = 10.75CYY371 pKa = 10.03GVSPSKK377 pKa = 11.02LIDD380 pKa = 3.28LCFTSVYY387 pKa = 10.97ADD389 pKa = 3.22TFLIRR394 pKa = 11.84FSEE397 pKa = 4.39VRR399 pKa = 11.84QIAPGEE405 pKa = 4.15TGVIADD411 pKa = 4.42YY412 pKa = 10.83NYY414 pKa = 11.06KK415 pKa = 10.7LPDD418 pKa = 3.66EE419 pKa = 4.37FTGCVIAWNTANQDD433 pKa = 2.9RR434 pKa = 11.84GQYY437 pKa = 9.81YY438 pKa = 9.57YY439 pKa = 10.87RR440 pKa = 11.84SSRR443 pKa = 11.84KK444 pKa = 7.58TKK446 pKa = 10.3LKK448 pKa = 10.27PFEE451 pKa = 5.22RR452 pKa = 11.84DD453 pKa = 3.0LSSDD457 pKa = 3.51EE458 pKa = 4.18NGVRR462 pKa = 11.84TLSTYY467 pKa = 11.13DD468 pKa = 4.03FYY470 pKa = 11.17PSVPLEE476 pKa = 4.01YY477 pKa = 9.74QATRR481 pKa = 11.84VVVLSFEE488 pKa = 4.61LLNAPATVCGPKK500 pKa = 10.42LSTSLIKK507 pKa = 10.38NQCVNFNFNGLKK519 pKa = 9.11GTGVLTDD526 pKa = 3.51SSKK529 pKa = 11.0KK530 pKa = 9.96FQSFQQFGRR539 pKa = 11.84DD540 pKa = 3.18ASDD543 pKa = 3.34FTDD546 pKa = 3.68SVRR549 pKa = 11.84DD550 pKa = 3.75PQTLQILDD558 pKa = 3.94ISPCSFGGVSVITPGTNASSEE579 pKa = 4.47VAVLYY584 pKa = 10.73QDD586 pKa = 4.29VNCTDD591 pKa = 3.01VPTAIRR597 pKa = 11.84ADD599 pKa = 3.52QLTPAWRR606 pKa = 11.84VYY608 pKa = 10.25SAGVNVFQTQAGCLIGAEE626 pKa = 4.44HH627 pKa = 6.46VNASYY632 pKa = 11.03EE633 pKa = 4.01CDD635 pKa = 3.21IPIGAGICASYY646 pKa = 8.37HH647 pKa = 4.65TASLLRR653 pKa = 11.84NTGQKK658 pKa = 10.51SIVAYY663 pKa = 9.53TMSLGAEE670 pKa = 3.91NSIAYY675 pKa = 9.44ANNSIAIPTNFSISVTTEE693 pKa = 3.49VMPVSMAKK701 pKa = 9.66TSVDD705 pKa = 3.1CTMYY709 pKa = 10.34ICGDD713 pKa = 3.6SQEE716 pKa = 4.51CSNLLLQYY724 pKa = 11.24GSFCTQLNRR733 pKa = 11.84ALSGIAVEE741 pKa = 4.22QDD743 pKa = 2.97KK744 pKa = 8.86NTQEE748 pKa = 3.75VFAQVKK754 pKa = 9.11QMYY757 pKa = 7.06KK758 pKa = 9.37TPAIKK763 pKa = 10.61DD764 pKa = 3.33FGGFNFSQILPDD776 pKa = 3.66PSKK779 pKa = 8.47PTKK782 pKa = 10.11RR783 pKa = 11.84SFIEE787 pKa = 4.3DD788 pKa = 3.61LLFNKK793 pKa = 7.99VTLADD798 pKa = 3.57AGFMKK803 pKa = 10.37QYY805 pKa = 10.74GEE807 pKa = 4.27CLGDD811 pKa = 3.41ISARR815 pKa = 11.84DD816 pKa = 4.03LICAQKK822 pKa = 10.77FNGLTVLPPLLTDD835 pKa = 3.7EE836 pKa = 5.07MIAAYY841 pKa = 8.13TAALVSGTATAGWTFGAGAALQIPFAMQMAYY872 pKa = 10.08RR873 pKa = 11.84FNGIGVTQNVLYY885 pKa = 10.46EE886 pKa = 4.09NQKK889 pKa = 10.62QIANQFNKK897 pKa = 10.46AISQIQEE904 pKa = 4.19SLTTTSTALGKK915 pKa = 10.37LQDD918 pKa = 3.95VVNQNAQALNTLVKK932 pKa = 10.38QLSSNFGAISSVLNDD947 pKa = 3.17ILSRR951 pKa = 11.84LDD953 pKa = 3.31KK954 pKa = 11.45VEE956 pKa = 5.19AEE958 pKa = 4.28VQIDD962 pKa = 3.56RR963 pKa = 11.84LITGRR968 pKa = 11.84LQSLQTYY975 pKa = 7.4VTQQLIRR982 pKa = 11.84AAEE985 pKa = 3.85IRR987 pKa = 11.84ASANLAATKK996 pKa = 9.96MSEE999 pKa = 4.28CVLGQSKK1006 pKa = 10.12RR1007 pKa = 11.84VDD1009 pKa = 4.0FCGKK1013 pKa = 9.92GYY1015 pKa = 10.57HH1016 pKa = 6.84LMSFPQAAPHH1026 pKa = 5.76GVVFLHH1032 pKa = 5.25VTYY1035 pKa = 10.82VPSQEE1040 pKa = 4.9RR1041 pKa = 11.84NFTTAPAICHH1051 pKa = 5.3QGKK1054 pKa = 10.39AYY1056 pKa = 9.98FPRR1059 pKa = 11.84EE1060 pKa = 3.84GVFVSNGTSWFITQRR1075 pKa = 11.84NFFSPQIITTDD1086 pKa = 3.16NTFVSGNCDD1095 pKa = 3.2VVIGIINNTVYY1106 pKa = 10.97DD1107 pKa = 4.22PLQPEE1112 pKa = 4.19LDD1114 pKa = 3.85SFKK1117 pKa = 11.4EE1118 pKa = 3.91EE1119 pKa = 3.56LDD1121 pKa = 3.63KK1122 pKa = 11.53YY1123 pKa = 10.15FKK1125 pKa = 10.99NHH1127 pKa = 6.01TSPDD1131 pKa = 3.14VDD1133 pKa = 4.4LGDD1136 pKa = 3.31ISGINASVVNIQKK1149 pKa = 10.63EE1150 pKa = 3.92IDD1152 pKa = 3.67RR1153 pKa = 11.84LNEE1156 pKa = 3.5VAKK1159 pKa = 10.61NLNEE1163 pKa = 4.14SLIDD1167 pKa = 3.75LQEE1170 pKa = 4.17LGKK1173 pKa = 10.54YY1174 pKa = 6.98EE1175 pKa = 5.3QYY1177 pKa = 10.78IKK1179 pKa = 10.03WPWYY1183 pKa = 8.39VWLGFIAGLIAIVMATILLCCMTSCCSCLKK1213 pKa = 10.16GACSCGSCCKK1223 pKa = 10.16FDD1225 pKa = 4.43EE1226 pKa = 5.39DD1227 pKa = 5.38DD1228 pKa = 4.38SEE1230 pKa = 4.36PVLKK1234 pKa = 10.36GVKK1237 pKa = 9.35LHH1239 pKa = 5.3YY1240 pKa = 9.44TT1241 pKa = 3.44
Molecular weight: 137.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9QTB5|R9QTB5_SARS Membrane protein OS=Bat coronavirus Cp/Yunnan2011 OX=1283333 GN=M PE=3 SV=1
MM1 pKa = 7.79SDD3 pKa = 4.35NGPQQNQRR11 pKa = 11.84SAPRR15 pKa = 11.84ITFGGPTDD23 pKa = 4.07SADD26 pKa = 3.37NNQDD30 pKa = 3.01GGRR33 pKa = 11.84SGARR37 pKa = 11.84PKK39 pKa = 10.25QRR41 pKa = 11.84RR42 pKa = 11.84PQGLPNNTASWFTALTQHH60 pKa = 6.06GKK62 pKa = 10.36EE63 pKa = 3.96EE64 pKa = 4.07LRR66 pKa = 11.84FPRR69 pKa = 11.84GQGVPINTNSGKK81 pKa = 10.01DD82 pKa = 3.58DD83 pKa = 3.56QIGYY87 pKa = 8.17YY88 pKa = 9.92RR89 pKa = 11.84RR90 pKa = 11.84ATRR93 pKa = 11.84RR94 pKa = 11.84VRR96 pKa = 11.84GGDD99 pKa = 3.09GKK101 pKa = 10.39MKK103 pKa = 9.92EE104 pKa = 4.5LSPRR108 pKa = 11.84WYY110 pKa = 10.01FYY112 pKa = 11.5YY113 pKa = 10.73LGTGPEE119 pKa = 3.63ASLPYY124 pKa = 9.9GANKK128 pKa = 9.97EE129 pKa = 4.25GIVWVAIEE137 pKa = 4.45GALNTPKK144 pKa = 10.91DD145 pKa = 4.0HH146 pKa = 7.46IGTRR150 pKa = 11.84NPNNNAAIVLQLPQGTTLPKK170 pKa = 10.34GFYY173 pKa = 10.67AEE175 pKa = 4.5GSRR178 pKa = 11.84GGSQASSRR186 pKa = 11.84SSSRR190 pKa = 11.84SRR192 pKa = 11.84GNSRR196 pKa = 11.84NSTPGSSRR204 pKa = 11.84GNSPARR210 pKa = 11.84MASGGGEE217 pKa = 3.84TALALLLLDD226 pKa = 4.89RR227 pKa = 11.84LNQLEE232 pKa = 4.58SKK234 pKa = 10.86VSGKK238 pKa = 10.04GQQQQGQTVTKK249 pKa = 9.98KK250 pKa = 10.44SAAEE254 pKa = 3.73ASKK257 pKa = 10.76KK258 pKa = 9.61PRR260 pKa = 11.84QKK262 pKa = 9.66RR263 pKa = 11.84TATKK267 pKa = 10.12QYY269 pKa = 11.12NVTQAFGRR277 pKa = 11.84RR278 pKa = 11.84GPEE281 pKa = 3.4QTQGNFGDD289 pKa = 3.91QEE291 pKa = 4.91LIRR294 pKa = 11.84QGTDD298 pKa = 2.96YY299 pKa = 11.27KK300 pKa = 10.5HH301 pKa = 6.32WPQIAQFAPSASAFFGMSRR320 pKa = 11.84IGMEE324 pKa = 4.16VTPSGTWLTYY334 pKa = 10.42HH335 pKa = 7.06GAIKK339 pKa = 10.61LDD341 pKa = 4.18DD342 pKa = 4.89KK343 pKa = 11.64DD344 pKa = 3.91PQFKK348 pKa = 11.14DD349 pKa = 3.7NVILLNKK356 pKa = 10.01HH357 pKa = 4.67IDD359 pKa = 3.35AYY361 pKa = 9.77KK362 pKa = 9.88TFPPTEE368 pKa = 3.95PKK370 pKa = 9.88KK371 pKa = 10.6DD372 pKa = 3.48KK373 pKa = 10.73KK374 pKa = 10.99KK375 pKa = 9.88KK376 pKa = 8.29TDD378 pKa = 3.29EE379 pKa = 4.34AQPLPQRR386 pKa = 11.84QKK388 pKa = 10.35KK389 pKa = 8.88QPTVTLLPAADD400 pKa = 3.84MDD402 pKa = 4.26DD403 pKa = 5.3FSRR406 pKa = 11.84QLQNSMSGASADD418 pKa = 3.63STQAA422 pKa = 3.04
MM1 pKa = 7.79SDD3 pKa = 4.35NGPQQNQRR11 pKa = 11.84SAPRR15 pKa = 11.84ITFGGPTDD23 pKa = 4.07SADD26 pKa = 3.37NNQDD30 pKa = 3.01GGRR33 pKa = 11.84SGARR37 pKa = 11.84PKK39 pKa = 10.25QRR41 pKa = 11.84RR42 pKa = 11.84PQGLPNNTASWFTALTQHH60 pKa = 6.06GKK62 pKa = 10.36EE63 pKa = 3.96EE64 pKa = 4.07LRR66 pKa = 11.84FPRR69 pKa = 11.84GQGVPINTNSGKK81 pKa = 10.01DD82 pKa = 3.58DD83 pKa = 3.56QIGYY87 pKa = 8.17YY88 pKa = 9.92RR89 pKa = 11.84RR90 pKa = 11.84ATRR93 pKa = 11.84RR94 pKa = 11.84VRR96 pKa = 11.84GGDD99 pKa = 3.09GKK101 pKa = 10.39MKK103 pKa = 9.92EE104 pKa = 4.5LSPRR108 pKa = 11.84WYY110 pKa = 10.01FYY112 pKa = 11.5YY113 pKa = 10.73LGTGPEE119 pKa = 3.63ASLPYY124 pKa = 9.9GANKK128 pKa = 9.97EE129 pKa = 4.25GIVWVAIEE137 pKa = 4.45GALNTPKK144 pKa = 10.91DD145 pKa = 4.0HH146 pKa = 7.46IGTRR150 pKa = 11.84NPNNNAAIVLQLPQGTTLPKK170 pKa = 10.34GFYY173 pKa = 10.67AEE175 pKa = 4.5GSRR178 pKa = 11.84GGSQASSRR186 pKa = 11.84SSSRR190 pKa = 11.84SRR192 pKa = 11.84GNSRR196 pKa = 11.84NSTPGSSRR204 pKa = 11.84GNSPARR210 pKa = 11.84MASGGGEE217 pKa = 3.84TALALLLLDD226 pKa = 4.89RR227 pKa = 11.84LNQLEE232 pKa = 4.58SKK234 pKa = 10.86VSGKK238 pKa = 10.04GQQQQGQTVTKK249 pKa = 9.98KK250 pKa = 10.44SAAEE254 pKa = 3.73ASKK257 pKa = 10.76KK258 pKa = 9.61PRR260 pKa = 11.84QKK262 pKa = 9.66RR263 pKa = 11.84TATKK267 pKa = 10.12QYY269 pKa = 11.12NVTQAFGRR277 pKa = 11.84RR278 pKa = 11.84GPEE281 pKa = 3.4QTQGNFGDD289 pKa = 3.91QEE291 pKa = 4.91LIRR294 pKa = 11.84QGTDD298 pKa = 2.96YY299 pKa = 11.27KK300 pKa = 10.5HH301 pKa = 6.32WPQIAQFAPSASAFFGMSRR320 pKa = 11.84IGMEE324 pKa = 4.16VTPSGTWLTYY334 pKa = 10.42HH335 pKa = 7.06GAIKK339 pKa = 10.61LDD341 pKa = 4.18DD342 pKa = 4.89KK343 pKa = 11.64DD344 pKa = 3.91PQFKK348 pKa = 11.14DD349 pKa = 3.7NVILLNKK356 pKa = 10.01HH357 pKa = 4.67IDD359 pKa = 3.35AYY361 pKa = 9.77KK362 pKa = 9.88TFPPTEE368 pKa = 3.95PKK370 pKa = 9.88KK371 pKa = 10.6DD372 pKa = 3.48KK373 pKa = 10.73KK374 pKa = 10.99KK375 pKa = 9.88KK376 pKa = 8.29TDD378 pKa = 3.29EE379 pKa = 4.34AQPLPQRR386 pKa = 11.84QKK388 pKa = 10.35KK389 pKa = 8.88QPTVTLLPAADD400 pKa = 3.84MDD402 pKa = 4.26DD403 pKa = 5.3FSRR406 pKa = 11.84QLQNSMSGASADD418 pKa = 3.63STQAA422 pKa = 3.04
Molecular weight: 46.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
9245 |
76 |
4383 |
1320.7 |
147.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.226 ± 0.17 | 3.007 ± 0.429 |
5.343 ± 0.367 | 4.597 ± 0.583 |
4.878 ± 0.334 | 6.155 ± 0.567 |
2.001 ± 0.336 | 5.127 ± 0.357 |
5.635 ± 0.295 | 9.389 ± 0.639 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.326 ± 0.216 | 5.257 ± 0.306 |
4.024 ± 0.381 | 3.797 ± 0.712 |
3.797 ± 0.515 | 6.89 ± 0.548 |
6.977 ± 0.235 | 7.95 ± 0.718 |
1.136 ± 0.143 | 4.489 ± 0.387 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
