
Owenweeksia hongkongensis (strain DSM 17368 / CIP 108786 / JCM 12287 / NRRL B-23963 / UST20020801)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Schleiferiaceae; Owenweeksia; Owenweeksia hongkongensis
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
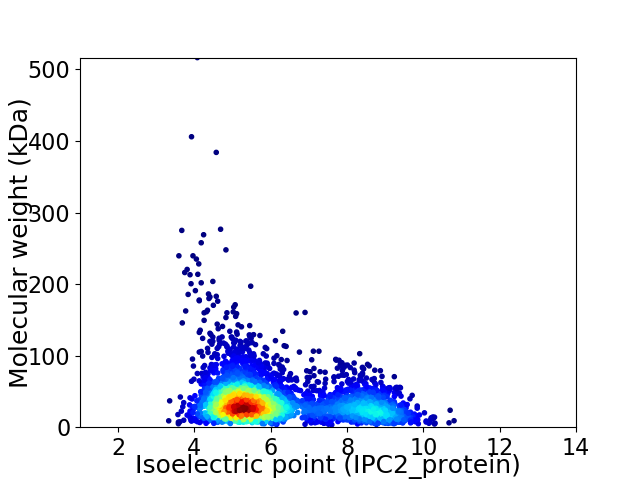
Virtual 2D-PAGE plot for 3471 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8R6A2|G8R6A2_OWEHD Uncharacterized protein OS=Owenweeksia hongkongensis (strain DSM 17368 / CIP 108786 / JCM 12287 / NRRL B-23963 / UST20020801) OX=926562 GN=Oweho_2351 PE=4 SV=1
MM1 pKa = 7.36NNYY4 pKa = 8.01TLSVNKK10 pKa = 10.0VSFAKK15 pKa = 10.86DD16 pKa = 3.11NGDD19 pKa = 3.03VSLAITLKK27 pKa = 10.84GSFPQPVNSIGNLVNDD43 pKa = 5.3TIQFDD48 pKa = 3.99LTSEE52 pKa = 4.09SGATVDD58 pKa = 3.45EE59 pKa = 4.59AVFTFQMGEE68 pKa = 4.26GYY70 pKa = 9.79PGDD73 pKa = 3.92FEE75 pKa = 5.54FINYY79 pKa = 7.0PQGYY83 pKa = 9.2GDD85 pKa = 4.23APINGDD91 pKa = 3.32VNADD95 pKa = 3.54NNEE98 pKa = 3.86EE99 pKa = 4.15CDD101 pKa = 4.93FEE103 pKa = 6.96LGDD106 pKa = 4.75CVWGVNPTGGLGSGEE121 pKa = 4.12GKK123 pKa = 10.16SLQVSVKK130 pKa = 9.58KK131 pKa = 10.8KK132 pKa = 10.16PGVTSYY138 pKa = 10.57EE139 pKa = 3.69ISSFLLTEE147 pKa = 4.16VEE149 pKa = 4.11ISGAMQLSAVMTLHH163 pKa = 6.97EE164 pKa = 4.33NAEE167 pKa = 4.24DD168 pKa = 3.69QLEE171 pKa = 4.37ATCEE175 pKa = 3.86EE176 pKa = 4.43LVNFTEE182 pKa = 4.89IGHH185 pKa = 7.02ALPIVTINEE194 pKa = 4.51SGDD197 pKa = 3.42STPRR201 pKa = 11.84SIGVIYY207 pKa = 9.17VNCC210 pKa = 4.27
MM1 pKa = 7.36NNYY4 pKa = 8.01TLSVNKK10 pKa = 10.0VSFAKK15 pKa = 10.86DD16 pKa = 3.11NGDD19 pKa = 3.03VSLAITLKK27 pKa = 10.84GSFPQPVNSIGNLVNDD43 pKa = 5.3TIQFDD48 pKa = 3.99LTSEE52 pKa = 4.09SGATVDD58 pKa = 3.45EE59 pKa = 4.59AVFTFQMGEE68 pKa = 4.26GYY70 pKa = 9.79PGDD73 pKa = 3.92FEE75 pKa = 5.54FINYY79 pKa = 7.0PQGYY83 pKa = 9.2GDD85 pKa = 4.23APINGDD91 pKa = 3.32VNADD95 pKa = 3.54NNEE98 pKa = 3.86EE99 pKa = 4.15CDD101 pKa = 4.93FEE103 pKa = 6.96LGDD106 pKa = 4.75CVWGVNPTGGLGSGEE121 pKa = 4.12GKK123 pKa = 10.16SLQVSVKK130 pKa = 9.58KK131 pKa = 10.8KK132 pKa = 10.16PGVTSYY138 pKa = 10.57EE139 pKa = 3.69ISSFLLTEE147 pKa = 4.16VEE149 pKa = 4.11ISGAMQLSAVMTLHH163 pKa = 6.97EE164 pKa = 4.33NAEE167 pKa = 4.24DD168 pKa = 3.69QLEE171 pKa = 4.37ATCEE175 pKa = 3.86EE176 pKa = 4.43LVNFTEE182 pKa = 4.89IGHH185 pKa = 7.02ALPIVTINEE194 pKa = 4.51SGDD197 pKa = 3.42STPRR201 pKa = 11.84SIGVIYY207 pKa = 9.17VNCC210 pKa = 4.27
Molecular weight: 22.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8R2M1|G8R2M1_OWEHD 50S ribosomal protein L15 OS=Owenweeksia hongkongensis (strain DSM 17368 / CIP 108786 / JCM 12287 / NRRL B-23963 / UST20020801) OX=926562 GN=rplO PE=3 SV=1
MM1 pKa = 7.61GVRR4 pKa = 11.84KK5 pKa = 9.94LNPVTPGQRR14 pKa = 11.84FRR16 pKa = 11.84IVNDD20 pKa = 2.91NSTITAAKK28 pKa = 9.34PEE30 pKa = 3.89KK31 pKa = 10.59SLVKK35 pKa = 10.44GKK37 pKa = 10.74SKK39 pKa = 10.99GGGRR43 pKa = 11.84NNTGKK48 pKa = 8.11MTIRR52 pKa = 11.84NVGGGHH58 pKa = 4.41KK59 pKa = 9.64QKK61 pKa = 10.77YY62 pKa = 8.52RR63 pKa = 11.84VVDD66 pKa = 4.69FKK68 pKa = 11.02RR69 pKa = 11.84DD70 pKa = 3.59KK71 pKa = 10.62EE72 pKa = 4.39GVPATVKK79 pKa = 10.53AVEE82 pKa = 4.0YY83 pKa = 10.59DD84 pKa = 3.47PNRR87 pKa = 11.84TARR90 pKa = 11.84IALLYY95 pKa = 10.03YY96 pKa = 10.51ADD98 pKa = 3.69GEE100 pKa = 4.31KK101 pKa = 10.29RR102 pKa = 11.84YY103 pKa = 9.45MIAPNGLQVGQTVVSGRR120 pKa = 11.84EE121 pKa = 3.68AAPEE125 pKa = 3.92IGNAMYY131 pKa = 10.77LSDD134 pKa = 4.36IPLGTVISCIEE145 pKa = 3.83MRR147 pKa = 11.84PGQGAVIARR156 pKa = 11.84SAGSFAQLAARR167 pKa = 11.84DD168 pKa = 3.75GKK170 pKa = 10.61YY171 pKa = 10.67AIIKK175 pKa = 9.48LPSGEE180 pKa = 3.93TRR182 pKa = 11.84MILITCMAMIGAVSNSDD199 pKa = 3.18NAQEE203 pKa = 4.26VSGKK207 pKa = 10.27AGRR210 pKa = 11.84KK211 pKa = 7.13RR212 pKa = 11.84WQGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84VRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.68HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.14GYY255 pKa = 7.49KK256 pKa = 8.5TRR258 pKa = 11.84APKK261 pKa = 10.32KK262 pKa = 9.2EE263 pKa = 3.77SSKK266 pKa = 11.08YY267 pKa = 9.4IIEE270 pKa = 4.11RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
MM1 pKa = 7.61GVRR4 pKa = 11.84KK5 pKa = 9.94LNPVTPGQRR14 pKa = 11.84FRR16 pKa = 11.84IVNDD20 pKa = 2.91NSTITAAKK28 pKa = 9.34PEE30 pKa = 3.89KK31 pKa = 10.59SLVKK35 pKa = 10.44GKK37 pKa = 10.74SKK39 pKa = 10.99GGGRR43 pKa = 11.84NNTGKK48 pKa = 8.11MTIRR52 pKa = 11.84NVGGGHH58 pKa = 4.41KK59 pKa = 9.64QKK61 pKa = 10.77YY62 pKa = 8.52RR63 pKa = 11.84VVDD66 pKa = 4.69FKK68 pKa = 11.02RR69 pKa = 11.84DD70 pKa = 3.59KK71 pKa = 10.62EE72 pKa = 4.39GVPATVKK79 pKa = 10.53AVEE82 pKa = 4.0YY83 pKa = 10.59DD84 pKa = 3.47PNRR87 pKa = 11.84TARR90 pKa = 11.84IALLYY95 pKa = 10.03YY96 pKa = 10.51ADD98 pKa = 3.69GEE100 pKa = 4.31KK101 pKa = 10.29RR102 pKa = 11.84YY103 pKa = 9.45MIAPNGLQVGQTVVSGRR120 pKa = 11.84EE121 pKa = 3.68AAPEE125 pKa = 3.92IGNAMYY131 pKa = 10.77LSDD134 pKa = 4.36IPLGTVISCIEE145 pKa = 3.83MRR147 pKa = 11.84PGQGAVIARR156 pKa = 11.84SAGSFAQLAARR167 pKa = 11.84DD168 pKa = 3.75GKK170 pKa = 10.61YY171 pKa = 10.67AIIKK175 pKa = 9.48LPSGEE180 pKa = 3.93TRR182 pKa = 11.84MILITCMAMIGAVSNSDD199 pKa = 3.18NAQEE203 pKa = 4.26VSGKK207 pKa = 10.27AGRR210 pKa = 11.84KK211 pKa = 7.13RR212 pKa = 11.84WQGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84VRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.68HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.13PRR245 pKa = 11.84SRR247 pKa = 11.84NGIPAKK253 pKa = 10.14GYY255 pKa = 7.49KK256 pKa = 8.5TRR258 pKa = 11.84APKK261 pKa = 10.32KK262 pKa = 9.2EE263 pKa = 3.77SSKK266 pKa = 11.08YY267 pKa = 9.4IIEE270 pKa = 4.11RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.1KK274 pKa = 9.82
Molecular weight: 29.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1212283 |
33 |
4841 |
349.3 |
39.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.896 ± 0.039 | 0.814 ± 0.015 |
5.697 ± 0.033 | 6.55 ± 0.059 |
4.986 ± 0.03 | 7.016 ± 0.047 |
1.843 ± 0.022 | 6.974 ± 0.034 |
6.612 ± 0.062 | 9.235 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.025 | 5.561 ± 0.038 |
3.725 ± 0.025 | 3.492 ± 0.025 |
3.692 ± 0.031 | 6.983 ± 0.051 |
5.796 ± 0.056 | 6.446 ± 0.034 |
1.196 ± 0.017 | 4.058 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
