
Xingshan nematode virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.04
Get precalculated fractions of proteins
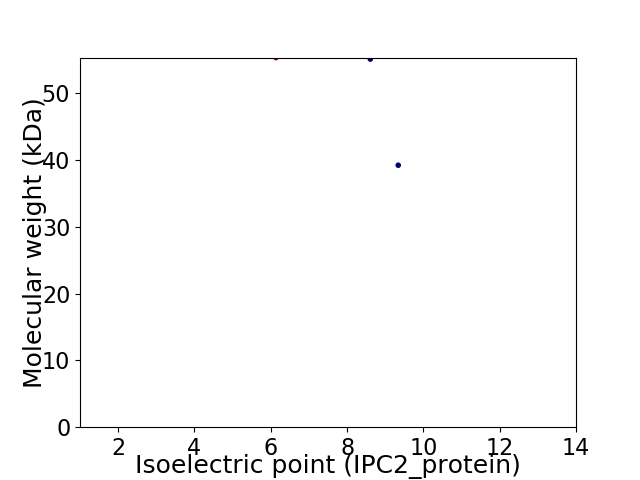
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KF94|A0A1L3KF94_9VIRU RNA-directed RNA polymerase OS=Xingshan nematode virus 5 OX=1923764 PE=4 SV=1
MM1 pKa = 7.52CPPWRR6 pKa = 11.84HH7 pKa = 5.43KK8 pKa = 10.56FSRR11 pKa = 11.84SPAGPRR17 pKa = 11.84ARR19 pKa = 11.84TQEE22 pKa = 4.35SATEE26 pKa = 4.07TPGFEE31 pKa = 4.24EE32 pKa = 4.54VGWSYY37 pKa = 11.47GVYY40 pKa = 10.36SRR42 pKa = 11.84MRR44 pKa = 11.84EE45 pKa = 3.91PTINPLMEE53 pKa = 4.28EE54 pKa = 3.96ALRR57 pKa = 11.84PYY59 pKa = 11.1AEE61 pKa = 4.46LFQYY65 pKa = 8.87TGQYY69 pKa = 9.32AWPPRR74 pKa = 11.84GPEE77 pKa = 3.76AEE79 pKa = 3.88RR80 pKa = 11.84VSLRR84 pKa = 11.84VHH86 pKa = 5.6ARR88 pKa = 11.84LRR90 pKa = 11.84AEE92 pKa = 4.22ILAGMQEE99 pKa = 4.05PTHH102 pKa = 7.83DD103 pKa = 3.72EE104 pKa = 4.39EE105 pKa = 4.47EE106 pKa = 4.22WLVAQTVKK114 pKa = 10.71AYY116 pKa = 10.46RR117 pKa = 11.84PARR120 pKa = 11.84WEE122 pKa = 3.85IPDD125 pKa = 5.45DD126 pKa = 3.91FLSWTHH132 pKa = 5.68FMRR135 pKa = 11.84VVARR139 pKa = 11.84LEE141 pKa = 4.46RR142 pKa = 11.84NSSPGWPYY150 pKa = 10.15CRR152 pKa = 11.84QYY154 pKa = 10.72GTLKK158 pKa = 10.63LWIGEE163 pKa = 4.02ILDD166 pKa = 4.08PDD168 pKa = 3.64EE169 pKa = 6.07AGIRR173 pKa = 11.84MLWDD177 pKa = 3.77LVQGRR182 pKa = 11.84LRR184 pKa = 11.84GDD186 pKa = 3.27EE187 pKa = 3.96AHH189 pKa = 7.79PIRR192 pKa = 11.84LFVKK196 pKa = 9.93PEE198 pKa = 3.49PHH200 pKa = 7.03KK201 pKa = 10.57ISKK204 pKa = 9.33LQEE207 pKa = 3.27GRR209 pKa = 11.84YY210 pKa = 9.66RR211 pKa = 11.84LISSVNVVDD220 pKa = 4.41QVIDD224 pKa = 3.35HH225 pKa = 6.56MLFDD229 pKa = 3.88MQNDD233 pKa = 3.73RR234 pKa = 11.84EE235 pKa = 4.46IAVHH239 pKa = 6.82LDD241 pKa = 3.15IPSKK245 pKa = 10.42PGWSPARR252 pKa = 11.84GGWRR256 pKa = 11.84MLQQFEE262 pKa = 4.56TAVSCDD268 pKa = 3.02KK269 pKa = 11.31SLWDD273 pKa = 3.95WGMPWWVARR282 pKa = 11.84LDD284 pKa = 3.92LKK286 pKa = 11.21VRR288 pKa = 11.84ASLCMNMQTAWMEE301 pKa = 3.92LAEE304 pKa = 4.04RR305 pKa = 11.84RR306 pKa = 11.84YY307 pKa = 10.3RR308 pKa = 11.84EE309 pKa = 4.19LYY311 pKa = 9.78QEE313 pKa = 4.15ARR315 pKa = 11.84FITSDD320 pKa = 3.46GQIFAQKK327 pKa = 9.57TGGLQKK333 pKa = 10.35SGCVNTISTNSRR345 pKa = 11.84GQYY348 pKa = 9.06LLHH351 pKa = 6.43MLACLRR357 pKa = 11.84SGEE360 pKa = 4.67DD361 pKa = 3.02HH362 pKa = 6.16TQCFWAMGDD371 pKa = 3.46DD372 pKa = 3.98TLQEE376 pKa = 4.11RR377 pKa = 11.84ASEE380 pKa = 4.52KK381 pKa = 10.68YY382 pKa = 9.95CAEE385 pKa = 4.09LGKK388 pKa = 10.39LCRR391 pKa = 11.84LKK393 pKa = 10.75EE394 pKa = 4.06VTPGIVFAGTRR405 pKa = 11.84LDD407 pKa = 3.59SEE409 pKa = 4.95GVHH412 pKa = 6.79PEE414 pKa = 4.13YY415 pKa = 10.51CSKK418 pKa = 10.91HH419 pKa = 5.66YY420 pKa = 8.08FTLLHH425 pKa = 6.64KK426 pKa = 10.54SQEE429 pKa = 4.28AVPDD433 pKa = 3.91TLSSYY438 pKa = 10.43VLLYY442 pKa = 11.0ARR444 pKa = 11.84DD445 pKa = 3.66QRR447 pKa = 11.84LEE449 pKa = 3.76ALLEE453 pKa = 4.02VARR456 pKa = 11.84RR457 pKa = 11.84LVPGVWRR464 pKa = 11.84SAVEE468 pKa = 3.61LRR470 pKa = 11.84GWYY473 pKa = 9.88DD474 pKa = 3.38GLIDD478 pKa = 3.96
MM1 pKa = 7.52CPPWRR6 pKa = 11.84HH7 pKa = 5.43KK8 pKa = 10.56FSRR11 pKa = 11.84SPAGPRR17 pKa = 11.84ARR19 pKa = 11.84TQEE22 pKa = 4.35SATEE26 pKa = 4.07TPGFEE31 pKa = 4.24EE32 pKa = 4.54VGWSYY37 pKa = 11.47GVYY40 pKa = 10.36SRR42 pKa = 11.84MRR44 pKa = 11.84EE45 pKa = 3.91PTINPLMEE53 pKa = 4.28EE54 pKa = 3.96ALRR57 pKa = 11.84PYY59 pKa = 11.1AEE61 pKa = 4.46LFQYY65 pKa = 8.87TGQYY69 pKa = 9.32AWPPRR74 pKa = 11.84GPEE77 pKa = 3.76AEE79 pKa = 3.88RR80 pKa = 11.84VSLRR84 pKa = 11.84VHH86 pKa = 5.6ARR88 pKa = 11.84LRR90 pKa = 11.84AEE92 pKa = 4.22ILAGMQEE99 pKa = 4.05PTHH102 pKa = 7.83DD103 pKa = 3.72EE104 pKa = 4.39EE105 pKa = 4.47EE106 pKa = 4.22WLVAQTVKK114 pKa = 10.71AYY116 pKa = 10.46RR117 pKa = 11.84PARR120 pKa = 11.84WEE122 pKa = 3.85IPDD125 pKa = 5.45DD126 pKa = 3.91FLSWTHH132 pKa = 5.68FMRR135 pKa = 11.84VVARR139 pKa = 11.84LEE141 pKa = 4.46RR142 pKa = 11.84NSSPGWPYY150 pKa = 10.15CRR152 pKa = 11.84QYY154 pKa = 10.72GTLKK158 pKa = 10.63LWIGEE163 pKa = 4.02ILDD166 pKa = 4.08PDD168 pKa = 3.64EE169 pKa = 6.07AGIRR173 pKa = 11.84MLWDD177 pKa = 3.77LVQGRR182 pKa = 11.84LRR184 pKa = 11.84GDD186 pKa = 3.27EE187 pKa = 3.96AHH189 pKa = 7.79PIRR192 pKa = 11.84LFVKK196 pKa = 9.93PEE198 pKa = 3.49PHH200 pKa = 7.03KK201 pKa = 10.57ISKK204 pKa = 9.33LQEE207 pKa = 3.27GRR209 pKa = 11.84YY210 pKa = 9.66RR211 pKa = 11.84LISSVNVVDD220 pKa = 4.41QVIDD224 pKa = 3.35HH225 pKa = 6.56MLFDD229 pKa = 3.88MQNDD233 pKa = 3.73RR234 pKa = 11.84EE235 pKa = 4.46IAVHH239 pKa = 6.82LDD241 pKa = 3.15IPSKK245 pKa = 10.42PGWSPARR252 pKa = 11.84GGWRR256 pKa = 11.84MLQQFEE262 pKa = 4.56TAVSCDD268 pKa = 3.02KK269 pKa = 11.31SLWDD273 pKa = 3.95WGMPWWVARR282 pKa = 11.84LDD284 pKa = 3.92LKK286 pKa = 11.21VRR288 pKa = 11.84ASLCMNMQTAWMEE301 pKa = 3.92LAEE304 pKa = 4.04RR305 pKa = 11.84RR306 pKa = 11.84YY307 pKa = 10.3RR308 pKa = 11.84EE309 pKa = 4.19LYY311 pKa = 9.78QEE313 pKa = 4.15ARR315 pKa = 11.84FITSDD320 pKa = 3.46GQIFAQKK327 pKa = 9.57TGGLQKK333 pKa = 10.35SGCVNTISTNSRR345 pKa = 11.84GQYY348 pKa = 9.06LLHH351 pKa = 6.43MLACLRR357 pKa = 11.84SGEE360 pKa = 4.67DD361 pKa = 3.02HH362 pKa = 6.16TQCFWAMGDD371 pKa = 3.46DD372 pKa = 3.98TLQEE376 pKa = 4.11RR377 pKa = 11.84ASEE380 pKa = 4.52KK381 pKa = 10.68YY382 pKa = 9.95CAEE385 pKa = 4.09LGKK388 pKa = 10.39LCRR391 pKa = 11.84LKK393 pKa = 10.75EE394 pKa = 4.06VTPGIVFAGTRR405 pKa = 11.84LDD407 pKa = 3.59SEE409 pKa = 4.95GVHH412 pKa = 6.79PEE414 pKa = 4.13YY415 pKa = 10.51CSKK418 pKa = 10.91HH419 pKa = 5.66YY420 pKa = 8.08FTLLHH425 pKa = 6.64KK426 pKa = 10.54SQEE429 pKa = 4.28AVPDD433 pKa = 3.91TLSSYY438 pKa = 10.43VLLYY442 pKa = 11.0ARR444 pKa = 11.84DD445 pKa = 3.66QRR447 pKa = 11.84LEE449 pKa = 3.76ALLEE453 pKa = 4.02VARR456 pKa = 11.84RR457 pKa = 11.84LVPGVWRR464 pKa = 11.84SAVEE468 pKa = 3.61LRR470 pKa = 11.84GWYY473 pKa = 9.88DD474 pKa = 3.38GLIDD478 pKa = 3.96
Molecular weight: 55.34 kDa
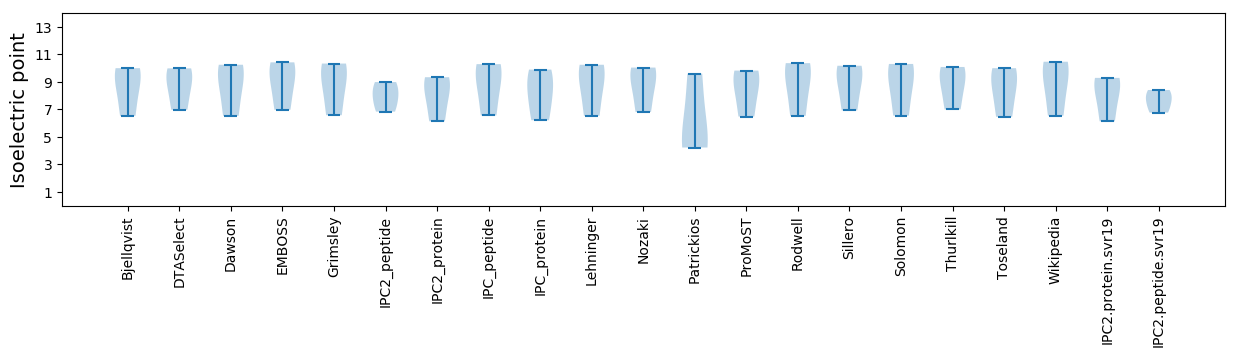
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF94|A0A1L3KF94_9VIRU RNA-directed RNA polymerase OS=Xingshan nematode virus 5 OX=1923764 PE=4 SV=1
MM1 pKa = 7.27STQQGNRR8 pKa = 11.84RR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84LRR14 pKa = 11.84NTVAALQQQLAQMQVQPVVQQTGGRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84AAAIAAAQVSTAPAATSLVVTTPPAPTLRR77 pKa = 11.84VTGTDD82 pKa = 3.06WLDD85 pKa = 3.58SEE87 pKa = 5.8AVGSMMQDD95 pKa = 2.86GWPVFEE101 pKa = 5.96LDD103 pKa = 4.57LNPTQFPGTHH113 pKa = 6.5LAALASGFEE122 pKa = 4.14KK123 pKa = 11.05YY124 pKa = 9.86RR125 pKa = 11.84FNRR128 pKa = 11.84LRR130 pKa = 11.84VLVSTRR136 pKa = 11.84APTTCGGGFIAGITPDD152 pKa = 3.38VYY154 pKa = 10.85QEE156 pKa = 4.03LVRR159 pKa = 11.84GIAGKK164 pKa = 10.3RR165 pKa = 11.84YY166 pKa = 9.01VRR168 pKa = 11.84SLQGAVSSSWWQPATLTMPCGVEE191 pKa = 3.73WLFTSPRR198 pKa = 11.84AEE200 pKa = 3.88RR201 pKa = 11.84SRR203 pKa = 11.84SSAGRR208 pKa = 11.84LFVVIDD214 pKa = 3.75GLPTVDD220 pKa = 5.0KK221 pKa = 11.23INVTLQLEE229 pKa = 4.28WDD231 pKa = 3.65ISLMAPAVPLEE242 pKa = 4.54HH243 pKa = 7.23KK244 pKa = 10.7SHH246 pKa = 6.86GDD248 pKa = 3.05WKK250 pKa = 10.72IPACTWVYY258 pKa = 11.09DD259 pKa = 3.69AAYY262 pKa = 9.17PNWASFKK269 pKa = 10.88AAGTPLDD276 pKa = 3.71EE277 pKa = 4.18WWNNSRR283 pKa = 11.84WSTLYY288 pKa = 10.58LVEE291 pKa = 4.64PGFMVGEE298 pKa = 4.21SPVRR302 pKa = 11.84VIIPHH307 pKa = 6.88DD308 pKa = 3.9FGGSDD313 pKa = 3.65HH314 pKa = 6.88GFLAYY319 pKa = 8.98STLYY323 pKa = 9.98AAKK326 pKa = 9.12TDD328 pKa = 3.56AAGTRR333 pKa = 11.84RR334 pKa = 11.84VKK336 pKa = 9.64VTANVDD342 pKa = 3.1VGPQHH347 pKa = 7.25IIEE350 pKa = 4.57IGPVNFF356 pKa = 4.46
MM1 pKa = 7.27STQQGNRR8 pKa = 11.84RR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84LRR14 pKa = 11.84NTVAALQQQLAQMQVQPVVQQTGGRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84AAAIAAAQVSTAPAATSLVVTTPPAPTLRR77 pKa = 11.84VTGTDD82 pKa = 3.06WLDD85 pKa = 3.58SEE87 pKa = 5.8AVGSMMQDD95 pKa = 2.86GWPVFEE101 pKa = 5.96LDD103 pKa = 4.57LNPTQFPGTHH113 pKa = 6.5LAALASGFEE122 pKa = 4.14KK123 pKa = 11.05YY124 pKa = 9.86RR125 pKa = 11.84FNRR128 pKa = 11.84LRR130 pKa = 11.84VLVSTRR136 pKa = 11.84APTTCGGGFIAGITPDD152 pKa = 3.38VYY154 pKa = 10.85QEE156 pKa = 4.03LVRR159 pKa = 11.84GIAGKK164 pKa = 10.3RR165 pKa = 11.84YY166 pKa = 9.01VRR168 pKa = 11.84SLQGAVSSSWWQPATLTMPCGVEE191 pKa = 3.73WLFTSPRR198 pKa = 11.84AEE200 pKa = 3.88RR201 pKa = 11.84SRR203 pKa = 11.84SSAGRR208 pKa = 11.84LFVVIDD214 pKa = 3.75GLPTVDD220 pKa = 5.0KK221 pKa = 11.23INVTLQLEE229 pKa = 4.28WDD231 pKa = 3.65ISLMAPAVPLEE242 pKa = 4.54HH243 pKa = 7.23KK244 pKa = 10.7SHH246 pKa = 6.86GDD248 pKa = 3.05WKK250 pKa = 10.72IPACTWVYY258 pKa = 11.09DD259 pKa = 3.69AAYY262 pKa = 9.17PNWASFKK269 pKa = 10.88AAGTPLDD276 pKa = 3.71EE277 pKa = 4.18WWNNSRR283 pKa = 11.84WSTLYY288 pKa = 10.58LVEE291 pKa = 4.64PGFMVGEE298 pKa = 4.21SPVRR302 pKa = 11.84VIIPHH307 pKa = 6.88DD308 pKa = 3.9FGGSDD313 pKa = 3.65HH314 pKa = 6.88GFLAYY319 pKa = 8.98STLYY323 pKa = 9.98AAKK326 pKa = 9.12TDD328 pKa = 3.56AAGTRR333 pKa = 11.84RR334 pKa = 11.84VKK336 pKa = 9.64VTANVDD342 pKa = 3.1VGPQHH347 pKa = 7.25IIEE350 pKa = 4.57IGPVNFF356 pKa = 4.46
Molecular weight: 39.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1326 |
356 |
492 |
442.0 |
49.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.824 ± 0.741 | 1.584 ± 0.268 |
4.223 ± 0.448 | 6.938 ± 1.136 |
3.092 ± 0.175 | 8.22 ± 0.943 |
2.036 ± 0.33 | 3.394 ± 0.168 |
3.922 ± 0.865 | 9.351 ± 0.562 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.941 ± 0.325 | 1.735 ± 0.35 |
6.184 ± 0.189 | 3.922 ± 0.723 |
8.974 ± 0.112 | 5.354 ± 0.728 |
5.204 ± 0.853 | 7.617 ± 0.827 |
3.017 ± 0.626 | 3.469 ± 0.399 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
