
Sphaerochaeta pleomorpha (strain ATCC BAA-1885 / DSM 22778 / Grapes)
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Sphaerochaetaceae; Sphaerochaeta; Sphaerochaeta pleomorpha
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
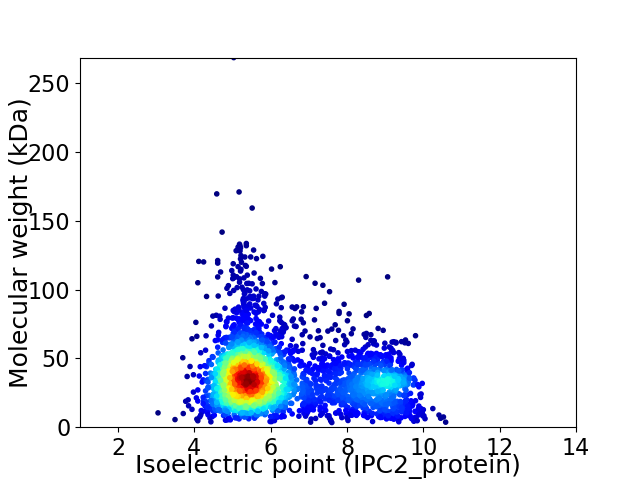
Virtual 2D-PAGE plot for 3150 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
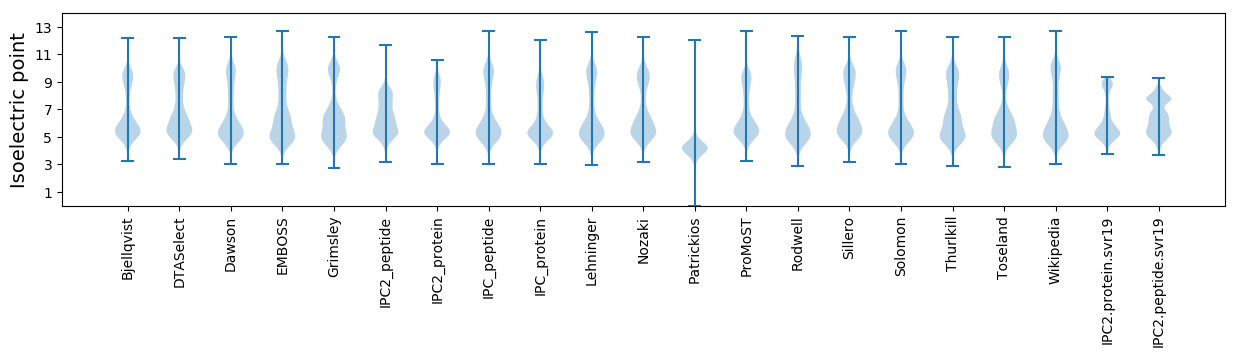
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G8QTW7|G8QTW7_SPHPG Adenylosuccinate synthetase OS=Sphaerochaeta pleomorpha (strain ATCC BAA-1885 / DSM 22778 / Grapes) OX=158190 GN=purA PE=3 SV=1
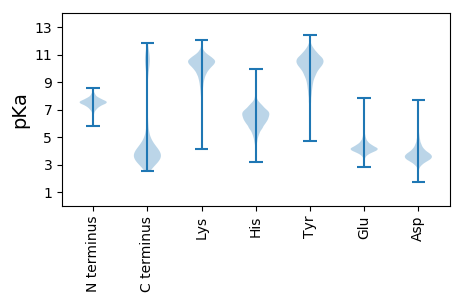
MM1 pKa = 7.45KK2 pKa = 10.4KK3 pKa = 10.27KK4 pKa = 9.79IALVMIVVMMVLFFTGCEE22 pKa = 3.72QQIPITEE29 pKa = 3.99ILSTVKK35 pKa = 10.57GYY37 pKa = 10.19SEE39 pKa = 5.47EE40 pKa = 3.92ILAMVASEE48 pKa = 4.8VEE50 pKa = 4.1TSLPDD55 pKa = 3.53SFTDD59 pKa = 3.52DD60 pKa = 3.21TLIAIDD66 pKa = 3.74TGISCRR72 pKa = 11.84VSNTYY77 pKa = 9.48TNEE80 pKa = 4.44LILDD84 pKa = 3.52ITLNNWVASDD94 pKa = 3.63GTVISGSIEE103 pKa = 3.6MDD105 pKa = 3.67LVYY108 pKa = 10.76DD109 pKa = 4.11ADD111 pKa = 3.53STYY114 pKa = 10.29IYY116 pKa = 9.88SIQSIVLMNYY126 pKa = 10.39DD127 pKa = 3.47LTSVYY132 pKa = 10.36FGEE135 pKa = 4.42EE136 pKa = 4.1VFDD139 pKa = 3.96NTPEE143 pKa = 3.85TAAFTFDD150 pKa = 3.46NATFQCLALSVDD162 pKa = 4.25GKK164 pKa = 10.03PLISNQLWLSLLTRR178 pKa = 11.84KK179 pKa = 9.53
MM1 pKa = 7.45KK2 pKa = 10.4KK3 pKa = 10.27KK4 pKa = 9.79IALVMIVVMMVLFFTGCEE22 pKa = 3.72QQIPITEE29 pKa = 3.99ILSTVKK35 pKa = 10.57GYY37 pKa = 10.19SEE39 pKa = 5.47EE40 pKa = 3.92ILAMVASEE48 pKa = 4.8VEE50 pKa = 4.1TSLPDD55 pKa = 3.53SFTDD59 pKa = 3.52DD60 pKa = 3.21TLIAIDD66 pKa = 3.74TGISCRR72 pKa = 11.84VSNTYY77 pKa = 9.48TNEE80 pKa = 4.44LILDD84 pKa = 3.52ITLNNWVASDD94 pKa = 3.63GTVISGSIEE103 pKa = 3.6MDD105 pKa = 3.67LVYY108 pKa = 10.76DD109 pKa = 4.11ADD111 pKa = 3.53STYY114 pKa = 10.29IYY116 pKa = 9.88SIQSIVLMNYY126 pKa = 10.39DD127 pKa = 3.47LTSVYY132 pKa = 10.36FGEE135 pKa = 4.42EE136 pKa = 4.1VFDD139 pKa = 3.96NTPEE143 pKa = 3.85TAAFTFDD150 pKa = 3.46NATFQCLALSVDD162 pKa = 4.25GKK164 pKa = 10.03PLISNQLWLSLLTRR178 pKa = 11.84KK179 pKa = 9.53
Molecular weight: 19.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8QQ36|G8QQ36_SPHPG Formyltetrahydrofolate deformylase OS=Sphaerochaeta pleomorpha (strain ATCC BAA-1885 / DSM 22778 / Grapes) OX=158190 GN=purU PE=3 SV=1
MM1 pKa = 6.35QTNTSARR8 pKa = 11.84RR9 pKa = 11.84GLLPALFIIPLLFFGLGFISILSALSAFVCMALPFILLAKK49 pKa = 9.87NKK51 pKa = 10.0RR52 pKa = 11.84KK53 pKa = 7.31TWCHH57 pKa = 5.94TYY59 pKa = 10.36CPRR62 pKa = 11.84ASLLQQTGKK71 pKa = 10.42KK72 pKa = 8.43GNKK75 pKa = 7.98MKK77 pKa = 10.35QAPRR81 pKa = 11.84QFTDD85 pKa = 3.65GSLRR89 pKa = 11.84KK90 pKa = 9.37ILLWYY95 pKa = 10.36FGLNLLFITGSTVRR109 pKa = 11.84IALGQMEE116 pKa = 4.82SMAFIRR122 pKa = 11.84LFIFLPLFPLPQLLDD137 pKa = 3.12IAAPPFLIHH146 pKa = 6.86LSYY149 pKa = 11.08RR150 pKa = 11.84LYY152 pKa = 11.77SMMLSSTILGVAFALIWRR170 pKa = 11.84NRR172 pKa = 11.84LWCAVCPVGNLSNKK186 pKa = 9.65ILGIKK191 pKa = 8.99NN192 pKa = 3.24
MM1 pKa = 6.35QTNTSARR8 pKa = 11.84RR9 pKa = 11.84GLLPALFIIPLLFFGLGFISILSALSAFVCMALPFILLAKK49 pKa = 9.87NKK51 pKa = 10.0RR52 pKa = 11.84KK53 pKa = 7.31TWCHH57 pKa = 5.94TYY59 pKa = 10.36CPRR62 pKa = 11.84ASLLQQTGKK71 pKa = 10.42KK72 pKa = 8.43GNKK75 pKa = 7.98MKK77 pKa = 10.35QAPRR81 pKa = 11.84QFTDD85 pKa = 3.65GSLRR89 pKa = 11.84KK90 pKa = 9.37ILLWYY95 pKa = 10.36FGLNLLFITGSTVRR109 pKa = 11.84IALGQMEE116 pKa = 4.82SMAFIRR122 pKa = 11.84LFIFLPLFPLPQLLDD137 pKa = 3.12IAAPPFLIHH146 pKa = 6.86LSYY149 pKa = 11.08RR150 pKa = 11.84LYY152 pKa = 11.77SMMLSSTILGVAFALIWRR170 pKa = 11.84NRR172 pKa = 11.84LWCAVCPVGNLSNKK186 pKa = 9.65ILGIKK191 pKa = 8.99NN192 pKa = 3.24
Molecular weight: 21.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1068925 |
29 |
2489 |
339.3 |
37.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.632 ± 0.047 | 1.388 ± 0.019 |
5.124 ± 0.032 | 6.162 ± 0.041 |
4.817 ± 0.03 | 7.195 ± 0.041 |
1.787 ± 0.018 | 7.125 ± 0.037 |
6.043 ± 0.03 | 10.465 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.769 ± 0.02 | 3.972 ± 0.025 |
3.83 ± 0.023 | 3.467 ± 0.024 |
4.184 ± 0.029 | 7.074 ± 0.037 |
5.566 ± 0.034 | 6.871 ± 0.036 |
1.021 ± 0.015 | 3.508 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
