
Sandfly fever Turkey virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Sicilian phlebovirus
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
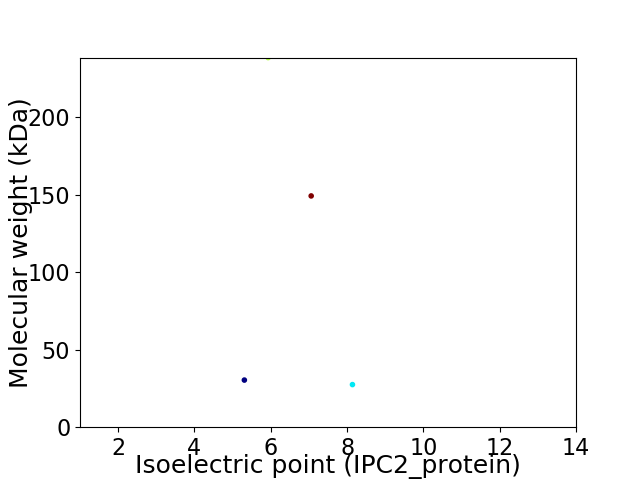
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D9YRN1|D9YRN1_9VIRU Polyprotein OS=Sandfly fever Turkey virus OX=688699 PE=4 SV=2
MM1 pKa = 7.61INNNMMNSQYY11 pKa = 10.34MFDD14 pKa = 3.88YY15 pKa = 10.01PAINIDD21 pKa = 3.83VRR23 pKa = 11.84CHH25 pKa = 6.41RR26 pKa = 11.84LLSSVSYY33 pKa = 9.13VAHH36 pKa = 6.45NKK38 pKa = 8.98FHH40 pKa = 5.86THH42 pKa = 6.73DD43 pKa = 3.32VSTYY47 pKa = 6.46EE48 pKa = 3.74HH49 pKa = 7.12CEE51 pKa = 3.64IPLEE55 pKa = 3.94KK56 pKa = 10.45LRR58 pKa = 11.84LGFGRR63 pKa = 11.84RR64 pKa = 11.84SSLSDD69 pKa = 3.92FYY71 pKa = 11.94SLGEE75 pKa = 4.57LPASWGPACYY85 pKa = 9.56ISSVKK90 pKa = 10.28PMVYY94 pKa = 9.27TFQGMASDD102 pKa = 4.59LSRR105 pKa = 11.84FDD107 pKa = 3.27LTSFSRR113 pKa = 11.84RR114 pKa = 11.84GLPNVLKK121 pKa = 10.71ALSWPLGIPDD131 pKa = 4.57CEE133 pKa = 4.11IFSICSDD140 pKa = 3.33RR141 pKa = 11.84FVRR144 pKa = 11.84GLQTRR149 pKa = 11.84DD150 pKa = 3.11QLMSYY155 pKa = 9.35ILRR158 pKa = 11.84MGDD161 pKa = 3.14SHH163 pKa = 8.69SLDD166 pKa = 3.4EE167 pKa = 5.98CIVQAHH173 pKa = 6.17KK174 pKa = 10.77KK175 pKa = 9.21ILQEE179 pKa = 3.78ARR181 pKa = 11.84RR182 pKa = 11.84LGLSDD187 pKa = 3.2EE188 pKa = 4.92HH189 pKa = 8.04YY190 pKa = 10.95NGYY193 pKa = 10.46DD194 pKa = 3.3LFRR197 pKa = 11.84EE198 pKa = 4.24IGSLVCLRR206 pKa = 11.84LINAEE211 pKa = 4.02PFDD214 pKa = 3.85TAFSGEE220 pKa = 4.08ALDD223 pKa = 3.34IRR225 pKa = 11.84TVIRR229 pKa = 11.84SYY231 pKa = 10.64RR232 pKa = 11.84ASDD235 pKa = 3.55PSASLTEE242 pKa = 4.23YY243 pKa = 11.08GNSLWTPIHH252 pKa = 5.65SHH254 pKa = 5.23VDD256 pKa = 3.44EE257 pKa = 4.83NDD259 pKa = 3.34EE260 pKa = 4.33SSSDD264 pKa = 3.37SDD266 pKa = 3.73FF267 pKa = 4.55
MM1 pKa = 7.61INNNMMNSQYY11 pKa = 10.34MFDD14 pKa = 3.88YY15 pKa = 10.01PAINIDD21 pKa = 3.83VRR23 pKa = 11.84CHH25 pKa = 6.41RR26 pKa = 11.84LLSSVSYY33 pKa = 9.13VAHH36 pKa = 6.45NKK38 pKa = 8.98FHH40 pKa = 5.86THH42 pKa = 6.73DD43 pKa = 3.32VSTYY47 pKa = 6.46EE48 pKa = 3.74HH49 pKa = 7.12CEE51 pKa = 3.64IPLEE55 pKa = 3.94KK56 pKa = 10.45LRR58 pKa = 11.84LGFGRR63 pKa = 11.84RR64 pKa = 11.84SSLSDD69 pKa = 3.92FYY71 pKa = 11.94SLGEE75 pKa = 4.57LPASWGPACYY85 pKa = 9.56ISSVKK90 pKa = 10.28PMVYY94 pKa = 9.27TFQGMASDD102 pKa = 4.59LSRR105 pKa = 11.84FDD107 pKa = 3.27LTSFSRR113 pKa = 11.84RR114 pKa = 11.84GLPNVLKK121 pKa = 10.71ALSWPLGIPDD131 pKa = 4.57CEE133 pKa = 4.11IFSICSDD140 pKa = 3.33RR141 pKa = 11.84FVRR144 pKa = 11.84GLQTRR149 pKa = 11.84DD150 pKa = 3.11QLMSYY155 pKa = 9.35ILRR158 pKa = 11.84MGDD161 pKa = 3.14SHH163 pKa = 8.69SLDD166 pKa = 3.4EE167 pKa = 5.98CIVQAHH173 pKa = 6.17KK174 pKa = 10.77KK175 pKa = 9.21ILQEE179 pKa = 3.78ARR181 pKa = 11.84RR182 pKa = 11.84LGLSDD187 pKa = 3.2EE188 pKa = 4.92HH189 pKa = 8.04YY190 pKa = 10.95NGYY193 pKa = 10.46DD194 pKa = 3.3LFRR197 pKa = 11.84EE198 pKa = 4.24IGSLVCLRR206 pKa = 11.84LINAEE211 pKa = 4.02PFDD214 pKa = 3.85TAFSGEE220 pKa = 4.08ALDD223 pKa = 3.34IRR225 pKa = 11.84TVIRR229 pKa = 11.84SYY231 pKa = 10.64RR232 pKa = 11.84ASDD235 pKa = 3.55PSASLTEE242 pKa = 4.23YY243 pKa = 11.08GNSLWTPIHH252 pKa = 5.65SHH254 pKa = 5.23VDD256 pKa = 3.44EE257 pKa = 4.83NDD259 pKa = 3.34EE260 pKa = 4.33SSSDD264 pKa = 3.37SDD266 pKa = 3.73FF267 pKa = 4.55
Molecular weight: 30.38 kDa
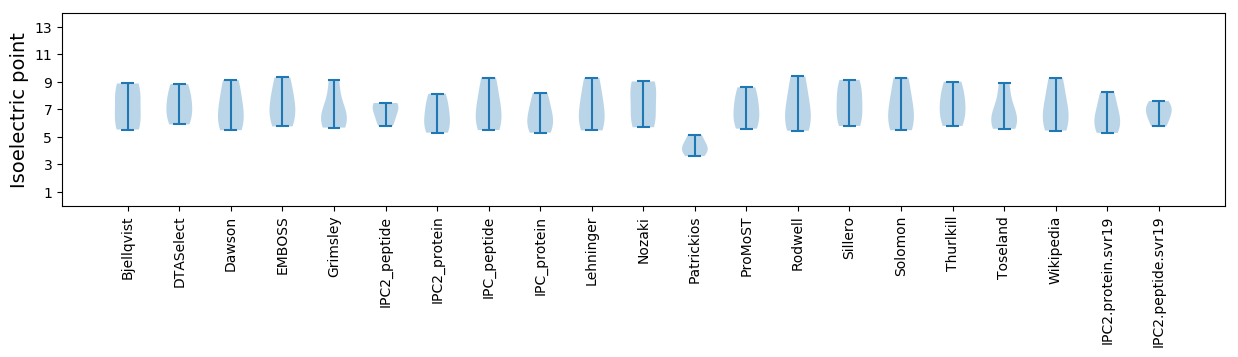
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F4Y5R8|F4Y5R8_9VIRU Nucleoprotein OS=Sandfly fever Turkey virus OX=688699 GN=N PE=3 SV=1
MM1 pKa = 8.37DD2 pKa = 5.38EE3 pKa = 4.07YY4 pKa = 11.49QKK6 pKa = 10.77IAVEE10 pKa = 4.37FGEE13 pKa = 4.12QAIDD17 pKa = 3.36EE18 pKa = 4.57AVIQDD23 pKa = 3.65WLQAFAYY30 pKa = 9.94QGFDD34 pKa = 3.03ARR36 pKa = 11.84TIIQNLVQLGGKK48 pKa = 8.02SWEE51 pKa = 3.93EE52 pKa = 3.77DD53 pKa = 3.16AKK55 pKa = 11.55KK56 pKa = 10.12MIILSLTRR64 pKa = 11.84GNKK67 pKa = 7.17PKK69 pKa = 10.89KK70 pKa = 8.29MVEE73 pKa = 3.87RR74 pKa = 11.84MSPEE78 pKa = 3.47GARR81 pKa = 11.84EE82 pKa = 4.1VKK84 pKa = 10.71SLVAKK89 pKa = 9.55YY90 pKa = 10.07KK91 pKa = 10.21IVEE94 pKa = 4.11GRR96 pKa = 11.84PGRR99 pKa = 11.84NGITLSRR106 pKa = 11.84VAAALAGWTVQAVEE120 pKa = 4.28VVEE123 pKa = 4.53NFLPVPGSTMDD134 pKa = 3.67RR135 pKa = 11.84MCGQTYY141 pKa = 8.66PRR143 pKa = 11.84QMMHH147 pKa = 6.99PSFAGLIDD155 pKa = 4.23PSLDD159 pKa = 3.36QEE161 pKa = 4.69DD162 pKa = 4.53FNAVLDD168 pKa = 3.77AHH170 pKa = 7.1KK171 pKa = 10.72LFLLMFSKK179 pKa = 9.88TINVSLRR186 pKa = 11.84GAQKK190 pKa = 10.49RR191 pKa = 11.84DD192 pKa = 3.17IEE194 pKa = 4.4EE195 pKa = 4.49SFSQPMLAAINSSFIDD211 pKa = 3.33NTQRR215 pKa = 11.84RR216 pKa = 11.84AFLTKK221 pKa = 10.63FGILTSGARR230 pKa = 11.84ATAVVKK236 pKa = 10.6KK237 pKa = 10.18IAEE240 pKa = 4.34AYY242 pKa = 8.93RR243 pKa = 11.84KK244 pKa = 10.29LEE246 pKa = 3.91
MM1 pKa = 8.37DD2 pKa = 5.38EE3 pKa = 4.07YY4 pKa = 11.49QKK6 pKa = 10.77IAVEE10 pKa = 4.37FGEE13 pKa = 4.12QAIDD17 pKa = 3.36EE18 pKa = 4.57AVIQDD23 pKa = 3.65WLQAFAYY30 pKa = 9.94QGFDD34 pKa = 3.03ARR36 pKa = 11.84TIIQNLVQLGGKK48 pKa = 8.02SWEE51 pKa = 3.93EE52 pKa = 3.77DD53 pKa = 3.16AKK55 pKa = 11.55KK56 pKa = 10.12MIILSLTRR64 pKa = 11.84GNKK67 pKa = 7.17PKK69 pKa = 10.89KK70 pKa = 8.29MVEE73 pKa = 3.87RR74 pKa = 11.84MSPEE78 pKa = 3.47GARR81 pKa = 11.84EE82 pKa = 4.1VKK84 pKa = 10.71SLVAKK89 pKa = 9.55YY90 pKa = 10.07KK91 pKa = 10.21IVEE94 pKa = 4.11GRR96 pKa = 11.84PGRR99 pKa = 11.84NGITLSRR106 pKa = 11.84VAAALAGWTVQAVEE120 pKa = 4.28VVEE123 pKa = 4.53NFLPVPGSTMDD134 pKa = 3.67RR135 pKa = 11.84MCGQTYY141 pKa = 8.66PRR143 pKa = 11.84QMMHH147 pKa = 6.99PSFAGLIDD155 pKa = 4.23PSLDD159 pKa = 3.36QEE161 pKa = 4.69DD162 pKa = 4.53FNAVLDD168 pKa = 3.77AHH170 pKa = 7.1KK171 pKa = 10.72LFLLMFSKK179 pKa = 9.88TINVSLRR186 pKa = 11.84GAQKK190 pKa = 10.49RR191 pKa = 11.84DD192 pKa = 3.17IEE194 pKa = 4.4EE195 pKa = 4.49SFSQPMLAAINSSFIDD211 pKa = 3.33NTQRR215 pKa = 11.84RR216 pKa = 11.84AFLTKK221 pKa = 10.63FGILTSGARR230 pKa = 11.84ATAVVKK236 pKa = 10.6KK237 pKa = 10.18IAEE240 pKa = 4.34AYY242 pKa = 8.93RR243 pKa = 11.84KK244 pKa = 10.29LEE246 pKa = 3.91
Molecular weight: 27.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3944 |
246 |
2090 |
986.0 |
111.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.882 ± 0.796 | 2.637 ± 0.789 |
5.806 ± 0.478 | 6.846 ± 0.231 |
4.615 ± 0.396 | 5.958 ± 0.106 |
2.561 ± 0.231 | 6.567 ± 0.125 |
6.237 ± 0.444 | 8.469 ± 0.379 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.22 ± 0.139 | 3.829 ± 0.121 |
3.448 ± 0.118 | 3.195 ± 0.383 |
5.958 ± 0.343 | 8.824 ± 0.49 |
5.325 ± 0.312 | 6.694 ± 0.371 |
1.141 ± 0.114 | 2.789 ± 0.181 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
