
Nitratifractor salsuginis (strain DSM 16511 / JCM 12458 / E9I37-1)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Campylobacterales; Sulfurovaceae; Nitratifractor; Nitratifractor salsuginis
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
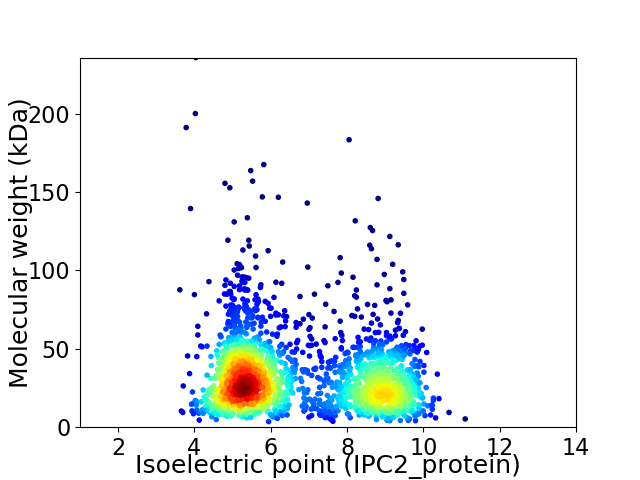
Virtual 2D-PAGE plot for 2076 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E6X004|E6X004_NITSE Histidine triad (HIT) protein OS=Nitratifractor salsuginis (strain DSM 16511 / JCM 12458 / E9I37-1) OX=749222 GN=Nitsa_1479 PE=4 SV=1
MM1 pKa = 7.22YY2 pKa = 10.12LLKK5 pKa = 10.67GALYY9 pKa = 10.38RR10 pKa = 11.84LFISGVIVLLSAISAQADD28 pKa = 2.86ISGIVFRR35 pKa = 11.84DD36 pKa = 3.87FNLNGVKK43 pKa = 9.36DD44 pKa = 3.79TLEE47 pKa = 4.37PGVAGLTVAAYY58 pKa = 9.81DD59 pKa = 4.34DD60 pKa = 4.04SGQAVDD66 pKa = 3.87TVMTNSDD73 pKa = 3.55GNYY76 pKa = 9.53TLATDD81 pKa = 4.22PGTYY85 pKa = 8.91RR86 pKa = 11.84VEE88 pKa = 4.28VSGVPAFLKK97 pKa = 10.57PGTSLSGNTRR107 pKa = 11.84PLVDD111 pKa = 3.9RR112 pKa = 11.84VTDD115 pKa = 4.06GATHH119 pKa = 5.88NVGLISAGEE128 pKa = 4.19YY129 pKa = 9.75CQSNPDD135 pKa = 3.27ILVTRR140 pKa = 11.84FTKK143 pKa = 10.58YY144 pKa = 10.64NRR146 pKa = 11.84DD147 pKa = 3.18GSNGQISVFLEE158 pKa = 3.73YY159 pKa = 10.35AYY161 pKa = 9.25TANTTDD167 pKa = 5.34DD168 pKa = 5.23DD169 pKa = 4.25STDD172 pKa = 3.52DD173 pKa = 4.03PKK175 pKa = 11.67AEE177 pKa = 4.51LGTYY181 pKa = 8.11SQIGSIYY188 pKa = 10.2GLAHH192 pKa = 6.49FKK194 pKa = 9.92EE195 pKa = 4.41ANITYY200 pKa = 10.22LGTYY204 pKa = 9.45LKK206 pKa = 10.12RR207 pKa = 11.84HH208 pKa = 6.16ADD210 pKa = 3.27IGPSGMGAIYY220 pKa = 10.01KK221 pKa = 9.65YY222 pKa = 10.62DD223 pKa = 3.63HH224 pKa = 7.14ASGTFSTLITLPTYY238 pKa = 10.46DD239 pKa = 4.05PRR241 pKa = 11.84NGAGQGYY248 pKa = 9.55DD249 pKa = 3.2WDD251 pKa = 4.89HH252 pKa = 6.6DD253 pKa = 3.94TEE255 pKa = 4.4AYY257 pKa = 9.86RR258 pKa = 11.84WVGKK262 pKa = 8.67TGIGDD267 pKa = 3.56IEE269 pKa = 4.19ISEE272 pKa = 4.19DD273 pKa = 3.56EE274 pKa = 4.11QRR276 pKa = 11.84LFAVDD281 pKa = 4.35LYY283 pKa = 11.33DD284 pKa = 4.04RR285 pKa = 11.84KK286 pKa = 10.91LYY288 pKa = 11.04VIDD291 pKa = 5.0LNQSGDD297 pKa = 3.69ATGHH301 pKa = 5.59TGYY304 pKa = 10.59DD305 pKa = 3.13IPNPCGNDD313 pKa = 2.83TDD315 pKa = 4.45FRR317 pKa = 11.84PMALGYY323 pKa = 9.99RR324 pKa = 11.84DD325 pKa = 3.14GKK327 pKa = 10.84LYY329 pKa = 11.24VGVTCTAEE337 pKa = 4.19STVDD341 pKa = 4.09PDD343 pKa = 4.2NQDD346 pKa = 3.5DD347 pKa = 4.17STNGPRR353 pKa = 11.84KK354 pKa = 9.59GDD356 pKa = 3.57RR357 pKa = 11.84NALSAHH363 pKa = 7.53IYY365 pKa = 10.56RR366 pKa = 11.84FDD368 pKa = 4.65PDD370 pKa = 3.64STSFEE375 pKa = 4.61PNAVLDD381 pKa = 3.69INLTYY386 pKa = 10.91DD387 pKa = 3.55RR388 pKa = 11.84GCIHH392 pKa = 7.03SSDD395 pKa = 4.58VSNQDD400 pKa = 3.17PSTCTYY406 pKa = 10.85QDD408 pKa = 3.46WDD410 pKa = 4.47DD411 pKa = 3.4QTQPYY416 pKa = 8.46RR417 pKa = 11.84ANWNPWQMDD426 pKa = 3.41YY427 pKa = 11.37DD428 pKa = 3.73IVFNDD433 pKa = 4.72KK434 pKa = 11.01KK435 pKa = 10.7PGDD438 pKa = 4.39IGNQDD443 pKa = 2.52NWIEE447 pKa = 3.91YY448 pKa = 7.25MQPLLSDD455 pKa = 3.39IVFDD459 pKa = 4.48NDD461 pKa = 2.78GSMIIQIRR469 pKa = 11.84DD470 pKa = 3.32VNGDD474 pKa = 2.87RR475 pKa = 11.84GGYY478 pKa = 9.62KK479 pKa = 10.16NYY481 pKa = 10.58SPNPNDD487 pKa = 3.42TTRR490 pKa = 11.84QNMNGEE496 pKa = 4.01GDD498 pKa = 3.4ILRR501 pKa = 11.84ACGNPQSGWTLEE513 pKa = 4.09NNGEE517 pKa = 4.27CGGVQTNGANNHH529 pKa = 6.0EE530 pKa = 4.53GPGGGEE536 pKa = 4.41YY537 pKa = 10.53YY538 pKa = 10.19WNDD541 pKa = 3.26QGPGGTNHH549 pKa = 5.51STAGSAYY556 pKa = 10.27QGDD559 pKa = 4.42AGHH562 pKa = 6.95SDD564 pKa = 3.15TSMGGLLVVPGYY576 pKa = 9.84PDD578 pKa = 3.5VVTGLMDD585 pKa = 3.3VTDD588 pKa = 4.05YY589 pKa = 11.71LNNGLGWLRR598 pKa = 11.84NDD600 pKa = 3.43TGEE603 pKa = 4.09LAKK606 pKa = 10.92DD607 pKa = 3.57GSGNPKK613 pKa = 9.9RR614 pKa = 11.84LLVSEE619 pKa = 4.91DD620 pKa = 4.14DD621 pKa = 3.29QDD623 pKa = 3.41KK624 pKa = 10.89FYY626 pKa = 11.42GKK628 pKa = 10.8GSGIGDD634 pKa = 4.4LEE636 pKa = 4.32ALCDD640 pKa = 3.73PAPIEE645 pKa = 3.83IGNYY649 pKa = 6.71VWEE652 pKa = 5.27DD653 pKa = 3.17SDD655 pKa = 4.92GDD657 pKa = 4.33GIQDD661 pKa = 3.79PGEE664 pKa = 4.1SPIANVTVKK673 pKa = 10.71LYY675 pKa = 10.5DD676 pKa = 3.59ANGQEE681 pKa = 4.44IGSTTTDD688 pKa = 2.99SNGEE692 pKa = 4.23YY693 pKa = 10.76YY694 pKa = 10.77FGGASNARR702 pKa = 11.84GVALAPHH709 pKa = 5.8THH711 pKa = 4.57YY712 pKa = 10.76QIRR715 pKa = 11.84IALNDD720 pKa = 3.05QHH722 pKa = 8.06LNGRR726 pKa = 11.84VPTTRR731 pKa = 11.84DD732 pKa = 3.58ANPSAPDD739 pKa = 3.53SDD741 pKa = 4.49DD742 pKa = 3.44QHH744 pKa = 8.9DD745 pKa = 3.84SDD747 pKa = 5.38GDD749 pKa = 3.76NGTLNPGFSTIDD761 pKa = 3.45YY762 pKa = 6.87TTGGAGKK769 pKa = 9.69NDD771 pKa = 3.2HH772 pKa = 6.42TLDD775 pKa = 3.68FGFVPPSSLGDD786 pKa = 3.89YY787 pKa = 10.52VWVDD791 pKa = 3.42SNRR794 pKa = 11.84DD795 pKa = 3.43GIQNDD800 pKa = 3.96GANAGIQGVTVHH812 pKa = 6.95LYY814 pKa = 10.81KK815 pKa = 10.85DD816 pKa = 3.7GQDD819 pKa = 3.25TGRR822 pKa = 11.84TATTDD827 pKa = 3.2GNGKK831 pKa = 9.28YY832 pKa = 10.46LFDD835 pKa = 3.86NLVPGSYY842 pKa = 9.48HH843 pKa = 6.8VKK845 pKa = 9.65FDD847 pKa = 3.55VPNGYY852 pKa = 9.3VVSPKK857 pKa = 10.66DD858 pKa = 3.24QGNDD862 pKa = 3.06EE863 pKa = 4.76TKK865 pKa = 11.02DD866 pKa = 3.46SDD868 pKa = 3.94ADD870 pKa = 3.76PATGATVDD878 pKa = 3.55TDD880 pKa = 4.04LSAGEE885 pKa = 5.22NDD887 pKa = 4.02LTWDD891 pKa = 3.23MGIYY895 pKa = 9.77PVADD899 pKa = 3.23VSLSKK904 pKa = 10.81DD905 pKa = 3.69VNQSTAYY912 pKa = 9.4EE913 pKa = 4.31GNRR916 pKa = 11.84VTFTITVHH924 pKa = 6.48NDD926 pKa = 3.26GPSVAHH932 pKa = 6.09NVTVTDD938 pKa = 4.39AVPDD942 pKa = 4.29GYY944 pKa = 11.38EE945 pKa = 4.01NVGDD949 pKa = 4.0ATNGASVSGSTVSFTVASLAVGADD973 pKa = 3.23ASFSFQADD981 pKa = 3.57YY982 pKa = 11.17KK983 pKa = 11.12EE984 pKa = 4.72SGEE987 pKa = 4.13HH988 pKa = 5.38TNWAEE993 pKa = 3.92VTAMDD998 pKa = 4.5EE999 pKa = 4.38ADD1001 pKa = 3.76VDD1003 pKa = 4.11SDD1005 pKa = 3.88PASDD1009 pKa = 3.87HH1010 pKa = 6.1TQDD1013 pKa = 5.21DD1014 pKa = 4.67LNDD1017 pKa = 4.18GQPDD1021 pKa = 3.91DD1022 pKa = 5.44DD1023 pKa = 4.64EE1024 pKa = 6.69SNATVSTGEE1033 pKa = 4.02KK1034 pKa = 9.13ATLGDD1039 pKa = 4.26YY1040 pKa = 10.81VWVDD1044 pKa = 3.42SNRR1047 pKa = 11.84DD1048 pKa = 3.43GIQNDD1053 pKa = 3.96GANAGIQGVTVHH1065 pKa = 6.95LYY1067 pKa = 10.81KK1068 pKa = 10.85DD1069 pKa = 3.7GQDD1072 pKa = 3.25TGRR1075 pKa = 11.84TATTDD1080 pKa = 3.29SNGKK1084 pKa = 9.18YY1085 pKa = 10.41LFDD1088 pKa = 3.77NLVPGSYY1095 pKa = 9.48HH1096 pKa = 6.8VKK1098 pKa = 9.65FDD1100 pKa = 3.55VPNGYY1105 pKa = 9.3VVSPKK1110 pKa = 10.66DD1111 pKa = 3.24QGNDD1115 pKa = 3.06EE1116 pKa = 4.76TKK1118 pKa = 11.02DD1119 pKa = 3.46SDD1121 pKa = 3.94ADD1123 pKa = 3.76PATGATVDD1131 pKa = 3.55TDD1133 pKa = 4.04LSAGEE1138 pKa = 5.22NDD1140 pKa = 4.02LTWDD1144 pKa = 3.23MGIYY1148 pKa = 9.83PVASQTASIGDD1159 pKa = 4.28YY1160 pKa = 11.13VWMDD1164 pKa = 3.82DD1165 pKa = 5.07NMNGLQDD1172 pKa = 4.89DD1173 pKa = 4.41NHH1175 pKa = 6.52PMSGVHH1181 pKa = 5.6VVLHH1185 pKa = 6.32RR1186 pKa = 11.84ADD1188 pKa = 3.49GSVVAEE1194 pKa = 4.14TDD1196 pKa = 3.76TNSSGQYY1203 pKa = 9.98YY1204 pKa = 10.36FGNLEE1209 pKa = 4.04AGDD1212 pKa = 4.4YY1213 pKa = 10.22YY1214 pKa = 11.71VSFDD1218 pKa = 3.44DD1219 pKa = 3.84QYY1221 pKa = 11.65YY1222 pKa = 8.0YY1223 pKa = 10.88TDD1225 pKa = 4.36PNVGSDD1231 pKa = 3.57DD1232 pKa = 4.49TIDD1235 pKa = 3.66SDD1237 pKa = 4.38VNRR1240 pKa = 11.84STFRR1244 pKa = 11.84TEE1246 pKa = 3.74TTHH1249 pKa = 7.13LDD1251 pKa = 2.82WGEE1254 pKa = 3.91RR1255 pKa = 11.84DD1256 pKa = 3.4MTIDD1260 pKa = 4.29AGITPTAHH1268 pKa = 6.24IGDD1271 pKa = 4.32YY1272 pKa = 10.73FWIDD1276 pKa = 3.32EE1277 pKa = 4.5NKK1279 pKa = 10.69NGIQDD1284 pKa = 3.75PGEE1287 pKa = 4.17PPVAGGGCRR1296 pKa = 11.84ALL1298 pKa = 3.86
MM1 pKa = 7.22YY2 pKa = 10.12LLKK5 pKa = 10.67GALYY9 pKa = 10.38RR10 pKa = 11.84LFISGVIVLLSAISAQADD28 pKa = 2.86ISGIVFRR35 pKa = 11.84DD36 pKa = 3.87FNLNGVKK43 pKa = 9.36DD44 pKa = 3.79TLEE47 pKa = 4.37PGVAGLTVAAYY58 pKa = 9.81DD59 pKa = 4.34DD60 pKa = 4.04SGQAVDD66 pKa = 3.87TVMTNSDD73 pKa = 3.55GNYY76 pKa = 9.53TLATDD81 pKa = 4.22PGTYY85 pKa = 8.91RR86 pKa = 11.84VEE88 pKa = 4.28VSGVPAFLKK97 pKa = 10.57PGTSLSGNTRR107 pKa = 11.84PLVDD111 pKa = 3.9RR112 pKa = 11.84VTDD115 pKa = 4.06GATHH119 pKa = 5.88NVGLISAGEE128 pKa = 4.19YY129 pKa = 9.75CQSNPDD135 pKa = 3.27ILVTRR140 pKa = 11.84FTKK143 pKa = 10.58YY144 pKa = 10.64NRR146 pKa = 11.84DD147 pKa = 3.18GSNGQISVFLEE158 pKa = 3.73YY159 pKa = 10.35AYY161 pKa = 9.25TANTTDD167 pKa = 5.34DD168 pKa = 5.23DD169 pKa = 4.25STDD172 pKa = 3.52DD173 pKa = 4.03PKK175 pKa = 11.67AEE177 pKa = 4.51LGTYY181 pKa = 8.11SQIGSIYY188 pKa = 10.2GLAHH192 pKa = 6.49FKK194 pKa = 9.92EE195 pKa = 4.41ANITYY200 pKa = 10.22LGTYY204 pKa = 9.45LKK206 pKa = 10.12RR207 pKa = 11.84HH208 pKa = 6.16ADD210 pKa = 3.27IGPSGMGAIYY220 pKa = 10.01KK221 pKa = 9.65YY222 pKa = 10.62DD223 pKa = 3.63HH224 pKa = 7.14ASGTFSTLITLPTYY238 pKa = 10.46DD239 pKa = 4.05PRR241 pKa = 11.84NGAGQGYY248 pKa = 9.55DD249 pKa = 3.2WDD251 pKa = 4.89HH252 pKa = 6.6DD253 pKa = 3.94TEE255 pKa = 4.4AYY257 pKa = 9.86RR258 pKa = 11.84WVGKK262 pKa = 8.67TGIGDD267 pKa = 3.56IEE269 pKa = 4.19ISEE272 pKa = 4.19DD273 pKa = 3.56EE274 pKa = 4.11QRR276 pKa = 11.84LFAVDD281 pKa = 4.35LYY283 pKa = 11.33DD284 pKa = 4.04RR285 pKa = 11.84KK286 pKa = 10.91LYY288 pKa = 11.04VIDD291 pKa = 5.0LNQSGDD297 pKa = 3.69ATGHH301 pKa = 5.59TGYY304 pKa = 10.59DD305 pKa = 3.13IPNPCGNDD313 pKa = 2.83TDD315 pKa = 4.45FRR317 pKa = 11.84PMALGYY323 pKa = 9.99RR324 pKa = 11.84DD325 pKa = 3.14GKK327 pKa = 10.84LYY329 pKa = 11.24VGVTCTAEE337 pKa = 4.19STVDD341 pKa = 4.09PDD343 pKa = 4.2NQDD346 pKa = 3.5DD347 pKa = 4.17STNGPRR353 pKa = 11.84KK354 pKa = 9.59GDD356 pKa = 3.57RR357 pKa = 11.84NALSAHH363 pKa = 7.53IYY365 pKa = 10.56RR366 pKa = 11.84FDD368 pKa = 4.65PDD370 pKa = 3.64STSFEE375 pKa = 4.61PNAVLDD381 pKa = 3.69INLTYY386 pKa = 10.91DD387 pKa = 3.55RR388 pKa = 11.84GCIHH392 pKa = 7.03SSDD395 pKa = 4.58VSNQDD400 pKa = 3.17PSTCTYY406 pKa = 10.85QDD408 pKa = 3.46WDD410 pKa = 4.47DD411 pKa = 3.4QTQPYY416 pKa = 8.46RR417 pKa = 11.84ANWNPWQMDD426 pKa = 3.41YY427 pKa = 11.37DD428 pKa = 3.73IVFNDD433 pKa = 4.72KK434 pKa = 11.01KK435 pKa = 10.7PGDD438 pKa = 4.39IGNQDD443 pKa = 2.52NWIEE447 pKa = 3.91YY448 pKa = 7.25MQPLLSDD455 pKa = 3.39IVFDD459 pKa = 4.48NDD461 pKa = 2.78GSMIIQIRR469 pKa = 11.84DD470 pKa = 3.32VNGDD474 pKa = 2.87RR475 pKa = 11.84GGYY478 pKa = 9.62KK479 pKa = 10.16NYY481 pKa = 10.58SPNPNDD487 pKa = 3.42TTRR490 pKa = 11.84QNMNGEE496 pKa = 4.01GDD498 pKa = 3.4ILRR501 pKa = 11.84ACGNPQSGWTLEE513 pKa = 4.09NNGEE517 pKa = 4.27CGGVQTNGANNHH529 pKa = 6.0EE530 pKa = 4.53GPGGGEE536 pKa = 4.41YY537 pKa = 10.53YY538 pKa = 10.19WNDD541 pKa = 3.26QGPGGTNHH549 pKa = 5.51STAGSAYY556 pKa = 10.27QGDD559 pKa = 4.42AGHH562 pKa = 6.95SDD564 pKa = 3.15TSMGGLLVVPGYY576 pKa = 9.84PDD578 pKa = 3.5VVTGLMDD585 pKa = 3.3VTDD588 pKa = 4.05YY589 pKa = 11.71LNNGLGWLRR598 pKa = 11.84NDD600 pKa = 3.43TGEE603 pKa = 4.09LAKK606 pKa = 10.92DD607 pKa = 3.57GSGNPKK613 pKa = 9.9RR614 pKa = 11.84LLVSEE619 pKa = 4.91DD620 pKa = 4.14DD621 pKa = 3.29QDD623 pKa = 3.41KK624 pKa = 10.89FYY626 pKa = 11.42GKK628 pKa = 10.8GSGIGDD634 pKa = 4.4LEE636 pKa = 4.32ALCDD640 pKa = 3.73PAPIEE645 pKa = 3.83IGNYY649 pKa = 6.71VWEE652 pKa = 5.27DD653 pKa = 3.17SDD655 pKa = 4.92GDD657 pKa = 4.33GIQDD661 pKa = 3.79PGEE664 pKa = 4.1SPIANVTVKK673 pKa = 10.71LYY675 pKa = 10.5DD676 pKa = 3.59ANGQEE681 pKa = 4.44IGSTTTDD688 pKa = 2.99SNGEE692 pKa = 4.23YY693 pKa = 10.76YY694 pKa = 10.77FGGASNARR702 pKa = 11.84GVALAPHH709 pKa = 5.8THH711 pKa = 4.57YY712 pKa = 10.76QIRR715 pKa = 11.84IALNDD720 pKa = 3.05QHH722 pKa = 8.06LNGRR726 pKa = 11.84VPTTRR731 pKa = 11.84DD732 pKa = 3.58ANPSAPDD739 pKa = 3.53SDD741 pKa = 4.49DD742 pKa = 3.44QHH744 pKa = 8.9DD745 pKa = 3.84SDD747 pKa = 5.38GDD749 pKa = 3.76NGTLNPGFSTIDD761 pKa = 3.45YY762 pKa = 6.87TTGGAGKK769 pKa = 9.69NDD771 pKa = 3.2HH772 pKa = 6.42TLDD775 pKa = 3.68FGFVPPSSLGDD786 pKa = 3.89YY787 pKa = 10.52VWVDD791 pKa = 3.42SNRR794 pKa = 11.84DD795 pKa = 3.43GIQNDD800 pKa = 3.96GANAGIQGVTVHH812 pKa = 6.95LYY814 pKa = 10.81KK815 pKa = 10.85DD816 pKa = 3.7GQDD819 pKa = 3.25TGRR822 pKa = 11.84TATTDD827 pKa = 3.2GNGKK831 pKa = 9.28YY832 pKa = 10.46LFDD835 pKa = 3.86NLVPGSYY842 pKa = 9.48HH843 pKa = 6.8VKK845 pKa = 9.65FDD847 pKa = 3.55VPNGYY852 pKa = 9.3VVSPKK857 pKa = 10.66DD858 pKa = 3.24QGNDD862 pKa = 3.06EE863 pKa = 4.76TKK865 pKa = 11.02DD866 pKa = 3.46SDD868 pKa = 3.94ADD870 pKa = 3.76PATGATVDD878 pKa = 3.55TDD880 pKa = 4.04LSAGEE885 pKa = 5.22NDD887 pKa = 4.02LTWDD891 pKa = 3.23MGIYY895 pKa = 9.77PVADD899 pKa = 3.23VSLSKK904 pKa = 10.81DD905 pKa = 3.69VNQSTAYY912 pKa = 9.4EE913 pKa = 4.31GNRR916 pKa = 11.84VTFTITVHH924 pKa = 6.48NDD926 pKa = 3.26GPSVAHH932 pKa = 6.09NVTVTDD938 pKa = 4.39AVPDD942 pKa = 4.29GYY944 pKa = 11.38EE945 pKa = 4.01NVGDD949 pKa = 4.0ATNGASVSGSTVSFTVASLAVGADD973 pKa = 3.23ASFSFQADD981 pKa = 3.57YY982 pKa = 11.17KK983 pKa = 11.12EE984 pKa = 4.72SGEE987 pKa = 4.13HH988 pKa = 5.38TNWAEE993 pKa = 3.92VTAMDD998 pKa = 4.5EE999 pKa = 4.38ADD1001 pKa = 3.76VDD1003 pKa = 4.11SDD1005 pKa = 3.88PASDD1009 pKa = 3.87HH1010 pKa = 6.1TQDD1013 pKa = 5.21DD1014 pKa = 4.67LNDD1017 pKa = 4.18GQPDD1021 pKa = 3.91DD1022 pKa = 5.44DD1023 pKa = 4.64EE1024 pKa = 6.69SNATVSTGEE1033 pKa = 4.02KK1034 pKa = 9.13ATLGDD1039 pKa = 4.26YY1040 pKa = 10.81VWVDD1044 pKa = 3.42SNRR1047 pKa = 11.84DD1048 pKa = 3.43GIQNDD1053 pKa = 3.96GANAGIQGVTVHH1065 pKa = 6.95LYY1067 pKa = 10.81KK1068 pKa = 10.85DD1069 pKa = 3.7GQDD1072 pKa = 3.25TGRR1075 pKa = 11.84TATTDD1080 pKa = 3.29SNGKK1084 pKa = 9.18YY1085 pKa = 10.41LFDD1088 pKa = 3.77NLVPGSYY1095 pKa = 9.48HH1096 pKa = 6.8VKK1098 pKa = 9.65FDD1100 pKa = 3.55VPNGYY1105 pKa = 9.3VVSPKK1110 pKa = 10.66DD1111 pKa = 3.24QGNDD1115 pKa = 3.06EE1116 pKa = 4.76TKK1118 pKa = 11.02DD1119 pKa = 3.46SDD1121 pKa = 3.94ADD1123 pKa = 3.76PATGATVDD1131 pKa = 3.55TDD1133 pKa = 4.04LSAGEE1138 pKa = 5.22NDD1140 pKa = 4.02LTWDD1144 pKa = 3.23MGIYY1148 pKa = 9.83PVASQTASIGDD1159 pKa = 4.28YY1160 pKa = 11.13VWMDD1164 pKa = 3.82DD1165 pKa = 5.07NMNGLQDD1172 pKa = 4.89DD1173 pKa = 4.41NHH1175 pKa = 6.52PMSGVHH1181 pKa = 5.6VVLHH1185 pKa = 6.32RR1186 pKa = 11.84ADD1188 pKa = 3.49GSVVAEE1194 pKa = 4.14TDD1196 pKa = 3.76TNSSGQYY1203 pKa = 9.98YY1204 pKa = 10.36FGNLEE1209 pKa = 4.04AGDD1212 pKa = 4.4YY1213 pKa = 10.22YY1214 pKa = 11.71VSFDD1218 pKa = 3.44DD1219 pKa = 3.84QYY1221 pKa = 11.65YY1222 pKa = 8.0YY1223 pKa = 10.88TDD1225 pKa = 4.36PNVGSDD1231 pKa = 3.57DD1232 pKa = 4.49TIDD1235 pKa = 3.66SDD1237 pKa = 4.38VNRR1240 pKa = 11.84STFRR1244 pKa = 11.84TEE1246 pKa = 3.74TTHH1249 pKa = 7.13LDD1251 pKa = 2.82WGEE1254 pKa = 3.91RR1255 pKa = 11.84DD1256 pKa = 3.4MTIDD1260 pKa = 4.29AGITPTAHH1268 pKa = 6.24IGDD1271 pKa = 4.32YY1272 pKa = 10.73FWIDD1276 pKa = 3.32EE1277 pKa = 4.5NKK1279 pKa = 10.69NGIQDD1284 pKa = 3.75PGEE1287 pKa = 4.17PPVAGGGCRR1296 pKa = 11.84ALL1298 pKa = 3.86
Molecular weight: 139.51 kDa
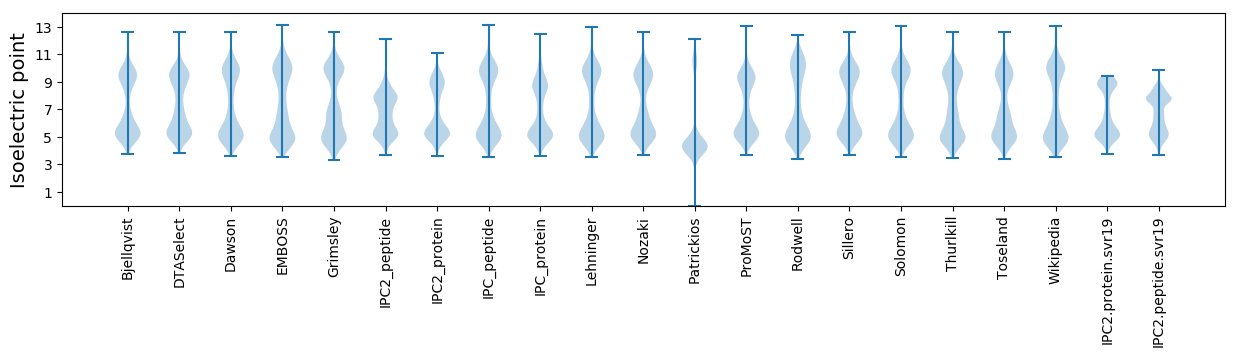
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E6WYY7|E6WYY7_NITSE Cysteine desulfuration protein SufE OS=Nitratifractor salsuginis (strain DSM 16511 / JCM 12458 / E9I37-1) OX=749222 GN=Nitsa_1322 PE=3 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.32RR14 pKa = 11.84THH16 pKa = 6.11GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.78TKK25 pKa = 10.46NGRR28 pKa = 11.84KK29 pKa = 9.3VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.86NTPRR12 pKa = 11.84KK13 pKa = 7.32RR14 pKa = 11.84THH16 pKa = 6.11GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.78TKK25 pKa = 10.46NGRR28 pKa = 11.84KK29 pKa = 9.3VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
630847 |
33 |
2213 |
303.9 |
34.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.281 ± 0.06 | 0.873 ± 0.021 |
5.365 ± 0.056 | 7.83 ± 0.083 |
4.298 ± 0.04 | 7.051 ± 0.056 |
2.202 ± 0.025 | 6.762 ± 0.051 |
6.422 ± 0.059 | 10.469 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.508 ± 0.029 | 3.403 ± 0.049 |
4.319 ± 0.036 | 2.894 ± 0.028 |
6.129 ± 0.068 | 5.402 ± 0.04 |
4.68 ± 0.04 | 6.308 ± 0.042 |
1.091 ± 0.022 | 3.713 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
