
Akodon montensis polyomavirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 7.11
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A343W7C6|A0A343W7C6_9POLY VP1 protein OS=Akodon montensis polyomavirus OX=2163422 PE=3 SV=1
MM1 pKa = 7.73ACQTQGRR8 pKa = 11.84KK9 pKa = 9.32RR10 pKa = 11.84LGPGKK15 pKa = 9.96APKK18 pKa = 10.01VAEE21 pKa = 3.87QRR23 pKa = 11.84LPRR26 pKa = 11.84ILRR29 pKa = 11.84KK30 pKa = 10.37GGVEE34 pKa = 3.96VLGTVPLSEE43 pKa = 3.94HH44 pKa = 4.0TQYY47 pKa = 10.96RR48 pKa = 11.84VEE50 pKa = 4.82CIVKK54 pKa = 9.43PVFGNEE60 pKa = 3.67PSNPPNYY67 pKa = 8.66WNHH70 pKa = 5.59SSALSGSALTDD81 pKa = 3.85GEE83 pKa = 4.57LHH85 pKa = 6.32LCYY88 pKa = 10.78SLMEE92 pKa = 4.13VQLPEE97 pKa = 3.9ISEE100 pKa = 4.18QCFEE104 pKa = 4.38DD105 pKa = 5.48AMIVWEE111 pKa = 4.43CFRR114 pKa = 11.84MEE116 pKa = 4.35TEE118 pKa = 4.63LLMTPKK124 pKa = 9.22ITVGGVNSSGGINSVQGTQMYY145 pKa = 8.32FWAVGGSPLDD155 pKa = 3.56VMYY158 pKa = 10.89LLPKK162 pKa = 10.36EE163 pKa = 4.11NMRR166 pKa = 11.84PRR168 pKa = 11.84GNLTAFDD175 pKa = 4.45SGHH178 pKa = 5.59SVYY181 pKa = 10.71DD182 pKa = 3.01IGVQKK187 pKa = 10.76GQVVNEE193 pKa = 4.26TFPVEE198 pKa = 4.05YY199 pKa = 9.75WVADD203 pKa = 3.64PSKK206 pKa = 11.08NDD208 pKa = 2.79NCRR211 pKa = 11.84YY212 pKa = 8.68FGRR215 pKa = 11.84VVGGAATPPVITYY228 pKa = 9.9SNSSTIPLLDD238 pKa = 3.81EE239 pKa = 4.58NGLGIMCNNGRR250 pKa = 11.84CYY252 pKa = 8.99ITCADD257 pKa = 3.16ITGYY261 pKa = 9.99LANRR265 pKa = 11.84VEE267 pKa = 4.41SSHH270 pKa = 6.24GRR272 pKa = 11.84FFRR275 pKa = 11.84LHH277 pKa = 5.05FRR279 pKa = 11.84QRR281 pKa = 11.84RR282 pKa = 11.84IKK284 pKa = 10.75NPYY287 pKa = 8.41TMNLLYY293 pKa = 10.69KK294 pKa = 10.33QVFQSNATAVEE305 pKa = 3.98AQKK308 pKa = 10.97EE309 pKa = 4.54VVEE312 pKa = 4.42VTTVEE317 pKa = 4.35DD318 pKa = 3.96EE319 pKa = 4.43SGLPMQVSNPRR330 pKa = 11.84CGQYY334 pKa = 9.99TPIQQQQASPLYY346 pKa = 9.66NVQGG350 pKa = 3.52
MM1 pKa = 7.73ACQTQGRR8 pKa = 11.84KK9 pKa = 9.32RR10 pKa = 11.84LGPGKK15 pKa = 9.96APKK18 pKa = 10.01VAEE21 pKa = 3.87QRR23 pKa = 11.84LPRR26 pKa = 11.84ILRR29 pKa = 11.84KK30 pKa = 10.37GGVEE34 pKa = 3.96VLGTVPLSEE43 pKa = 3.94HH44 pKa = 4.0TQYY47 pKa = 10.96RR48 pKa = 11.84VEE50 pKa = 4.82CIVKK54 pKa = 9.43PVFGNEE60 pKa = 3.67PSNPPNYY67 pKa = 8.66WNHH70 pKa = 5.59SSALSGSALTDD81 pKa = 3.85GEE83 pKa = 4.57LHH85 pKa = 6.32LCYY88 pKa = 10.78SLMEE92 pKa = 4.13VQLPEE97 pKa = 3.9ISEE100 pKa = 4.18QCFEE104 pKa = 4.38DD105 pKa = 5.48AMIVWEE111 pKa = 4.43CFRR114 pKa = 11.84MEE116 pKa = 4.35TEE118 pKa = 4.63LLMTPKK124 pKa = 9.22ITVGGVNSSGGINSVQGTQMYY145 pKa = 8.32FWAVGGSPLDD155 pKa = 3.56VMYY158 pKa = 10.89LLPKK162 pKa = 10.36EE163 pKa = 4.11NMRR166 pKa = 11.84PRR168 pKa = 11.84GNLTAFDD175 pKa = 4.45SGHH178 pKa = 5.59SVYY181 pKa = 10.71DD182 pKa = 3.01IGVQKK187 pKa = 10.76GQVVNEE193 pKa = 4.26TFPVEE198 pKa = 4.05YY199 pKa = 9.75WVADD203 pKa = 3.64PSKK206 pKa = 11.08NDD208 pKa = 2.79NCRR211 pKa = 11.84YY212 pKa = 8.68FGRR215 pKa = 11.84VVGGAATPPVITYY228 pKa = 9.9SNSSTIPLLDD238 pKa = 3.81EE239 pKa = 4.58NGLGIMCNNGRR250 pKa = 11.84CYY252 pKa = 8.99ITCADD257 pKa = 3.16ITGYY261 pKa = 9.99LANRR265 pKa = 11.84VEE267 pKa = 4.41SSHH270 pKa = 6.24GRR272 pKa = 11.84FFRR275 pKa = 11.84LHH277 pKa = 5.05FRR279 pKa = 11.84QRR281 pKa = 11.84RR282 pKa = 11.84IKK284 pKa = 10.75NPYY287 pKa = 8.41TMNLLYY293 pKa = 10.69KK294 pKa = 10.33QVFQSNATAVEE305 pKa = 3.98AQKK308 pKa = 10.97EE309 pKa = 4.54VVEE312 pKa = 4.42VTTVEE317 pKa = 4.35DD318 pKa = 3.96EE319 pKa = 4.43SGLPMQVSNPRR330 pKa = 11.84CGQYY334 pKa = 9.99TPIQQQQASPLYY346 pKa = 9.66NVQGG350 pKa = 3.52
Molecular weight: 38.86 kDa
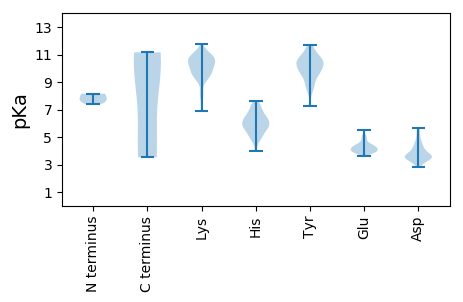
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A343W7C6|A0A343W7C6_9POLY VP1 protein OS=Akodon montensis polyomavirus OX=2163422 PE=3 SV=1
MM1 pKa = 7.72ALQVLPEE8 pKa = 4.1VPLGGGIPGVPDD20 pKa = 3.45WLFDD24 pKa = 4.46IIPAGLSTLQEE35 pKa = 4.11VFHH38 pKa = 7.15RR39 pKa = 11.84IATGIITSYY48 pKa = 9.54YY49 pKa = 7.22HH50 pKa = 5.81TGRR53 pKa = 11.84AIVRR57 pKa = 11.84RR58 pKa = 11.84AISVEE63 pKa = 3.94MQRR66 pKa = 11.84LLSDD70 pKa = 3.46LRR72 pKa = 11.84DD73 pKa = 3.65GFEE76 pKa = 4.21TTFEE80 pKa = 4.22ALGQGDD86 pKa = 4.33PVEE89 pKa = 5.13AIIDD93 pKa = 3.78QVRR96 pKa = 11.84AAHH99 pKa = 5.61STMIARR105 pKa = 11.84EE106 pKa = 3.76RR107 pKa = 11.84RR108 pKa = 11.84LIEE111 pKa = 3.83QGTPLLPNLGGQIVDD126 pKa = 4.15TVAGAAQTVSNIVVDD141 pKa = 3.79VTNIPMDD148 pKa = 4.66GYY150 pKa = 11.33NALQDD155 pKa = 3.65GVHH158 pKa = 7.27RR159 pKa = 11.84LGQWVQMAGSDD170 pKa = 3.46GGTPHH175 pKa = 6.71YY176 pKa = 10.62SYY178 pKa = 10.53PPWVLYY184 pKa = 9.94VLQEE188 pKa = 4.3LEE190 pKa = 4.4TPGPNISAASRR201 pKa = 11.84QPRR204 pKa = 11.84TLKK207 pKa = 10.6RR208 pKa = 11.84KK209 pKa = 9.52SQDD212 pKa = 2.88GLSNPRR218 pKa = 11.84QKK220 pKa = 10.63KK221 pKa = 9.25ARR223 pKa = 11.84TRR225 pKa = 11.84KK226 pKa = 7.93STQSGRR232 pKa = 11.84TKK234 pKa = 7.15TTKK237 pKa = 10.3NIKK240 pKa = 9.14KK241 pKa = 9.66GRR243 pKa = 11.84RR244 pKa = 11.84GGLRR248 pKa = 11.84HH249 pKa = 6.24SATKK253 pKa = 10.47
MM1 pKa = 7.72ALQVLPEE8 pKa = 4.1VPLGGGIPGVPDD20 pKa = 3.45WLFDD24 pKa = 4.46IIPAGLSTLQEE35 pKa = 4.11VFHH38 pKa = 7.15RR39 pKa = 11.84IATGIITSYY48 pKa = 9.54YY49 pKa = 7.22HH50 pKa = 5.81TGRR53 pKa = 11.84AIVRR57 pKa = 11.84RR58 pKa = 11.84AISVEE63 pKa = 3.94MQRR66 pKa = 11.84LLSDD70 pKa = 3.46LRR72 pKa = 11.84DD73 pKa = 3.65GFEE76 pKa = 4.21TTFEE80 pKa = 4.22ALGQGDD86 pKa = 4.33PVEE89 pKa = 5.13AIIDD93 pKa = 3.78QVRR96 pKa = 11.84AAHH99 pKa = 5.61STMIARR105 pKa = 11.84EE106 pKa = 3.76RR107 pKa = 11.84RR108 pKa = 11.84LIEE111 pKa = 3.83QGTPLLPNLGGQIVDD126 pKa = 4.15TVAGAAQTVSNIVVDD141 pKa = 3.79VTNIPMDD148 pKa = 4.66GYY150 pKa = 11.33NALQDD155 pKa = 3.65GVHH158 pKa = 7.27RR159 pKa = 11.84LGQWVQMAGSDD170 pKa = 3.46GGTPHH175 pKa = 6.71YY176 pKa = 10.62SYY178 pKa = 10.53PPWVLYY184 pKa = 9.94VLQEE188 pKa = 4.3LEE190 pKa = 4.4TPGPNISAASRR201 pKa = 11.84QPRR204 pKa = 11.84TLKK207 pKa = 10.6RR208 pKa = 11.84KK209 pKa = 9.52SQDD212 pKa = 2.88GLSNPRR218 pKa = 11.84QKK220 pKa = 10.63KK221 pKa = 9.25ARR223 pKa = 11.84TRR225 pKa = 11.84KK226 pKa = 7.93STQSGRR232 pKa = 11.84TKK234 pKa = 7.15TTKK237 pKa = 10.3NIKK240 pKa = 9.14KK241 pKa = 9.66GRR243 pKa = 11.84RR244 pKa = 11.84GGLRR248 pKa = 11.84HH249 pKa = 6.24SATKK253 pKa = 10.47
Molecular weight: 27.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1852 |
186 |
681 |
370.4 |
41.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.263 ± 1.71 | 2.7 ± 1.05 |
4.644 ± 0.824 | 6.371 ± 0.592 |
2.808 ± 0.587 | 8.207 ± 1.829 |
2.484 ± 0.283 | 5.022 ± 0.528 |
6.21 ± 1.679 | 9.017 ± 0.558 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.808 ± 0.397 | 4.752 ± 0.682 |
5.724 ± 0.303 | 5.022 ± 0.523 |
5.508 ± 0.616 | 5.616 ± 0.405 |
5.13 ± 0.888 | 6.695 ± 0.702 |
1.404 ± 0.264 | 3.618 ± 0.574 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
