
Canine distemper virus (strain Onderstepoort) (CDV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Morbillivirus; Canine morbillivirus
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
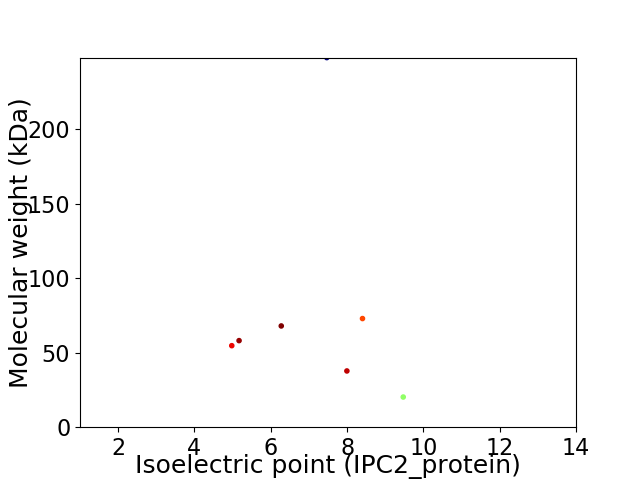
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
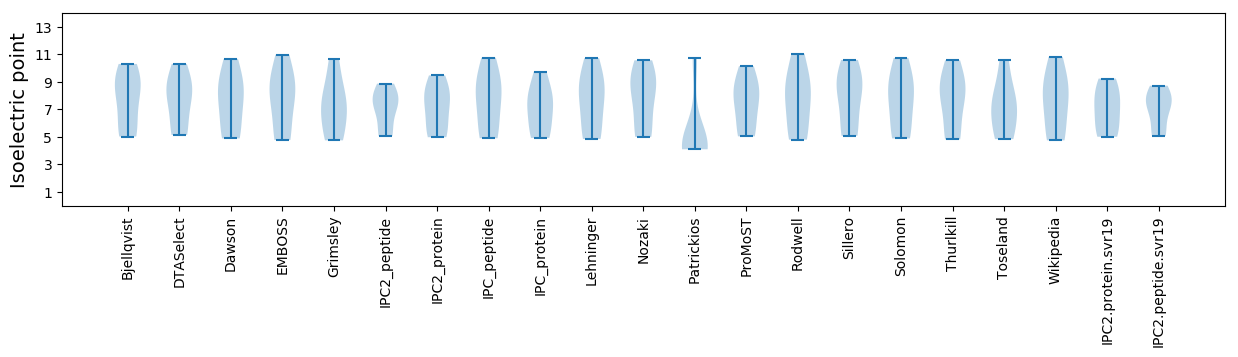
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O89326|O89326_CDVO Matrix protein OS=Canine distemper virus (strain Onderstepoort) OX=11233 PE=4 SV=1
MM1 pKa = 8.2AEE3 pKa = 3.69EE4 pKa = 3.84QAYY7 pKa = 9.61HH8 pKa = 5.38VSKK11 pKa = 10.9GLEE14 pKa = 3.92CLKK17 pKa = 10.59ALRR20 pKa = 11.84EE21 pKa = 4.01NPPDD25 pKa = 3.3IEE27 pKa = 4.62EE28 pKa = 4.1IQEE31 pKa = 4.01VSSLRR36 pKa = 11.84DD37 pKa = 3.28QTCNPGQEE45 pKa = 4.05NGTTGMQEE53 pKa = 4.16EE54 pKa = 4.73EE55 pKa = 4.27DD56 pKa = 3.95SQNLDD61 pKa = 3.42EE62 pKa = 4.54SHH64 pKa = 6.74EE65 pKa = 4.09PTKK68 pKa = 10.71GSNYY72 pKa = 9.5VGHH75 pKa = 6.68VPQNNPGCGEE85 pKa = 4.14RR86 pKa = 11.84NTALVEE92 pKa = 4.02AEE94 pKa = 4.06RR95 pKa = 11.84PPRR98 pKa = 11.84EE99 pKa = 4.69DD100 pKa = 3.04IQPGPGIRR108 pKa = 11.84CDD110 pKa = 3.25HH111 pKa = 6.72VYY113 pKa = 10.8DD114 pKa = 4.13HH115 pKa = 7.06SGEE118 pKa = 4.14EE119 pKa = 3.94VKK121 pKa = 11.05GIEE124 pKa = 4.43DD125 pKa = 3.86ADD127 pKa = 4.13SLVVPAGTVGNRR139 pKa = 11.84GFEE142 pKa = 4.04RR143 pKa = 11.84GEE145 pKa = 4.06GSLDD149 pKa = 4.26DD150 pKa = 3.79STEE153 pKa = 4.11DD154 pKa = 3.24SGEE157 pKa = 4.26DD158 pKa = 3.59YY159 pKa = 11.42SEE161 pKa = 4.91GNASSNWGYY170 pKa = 11.52SFGLKK175 pKa = 9.29PDD177 pKa = 3.53RR178 pKa = 11.84AADD181 pKa = 3.34VSMLMEE187 pKa = 4.64EE188 pKa = 4.45EE189 pKa = 4.38LSALLRR195 pKa = 11.84TSRR198 pKa = 11.84NVGIQKK204 pKa = 10.0RR205 pKa = 11.84DD206 pKa = 3.17GKK208 pKa = 8.63TLQFPHH214 pKa = 6.76NPEE217 pKa = 3.84GKK219 pKa = 8.16TRR221 pKa = 11.84DD222 pKa = 4.04PEE224 pKa = 4.44CGSIKK229 pKa = 10.48KK230 pKa = 8.55GTEE233 pKa = 3.79EE234 pKa = 4.29RR235 pKa = 11.84SVSHH239 pKa = 6.08GMGIVAGSTSGATQSALKK257 pKa = 9.2STGGSSEE264 pKa = 4.18PSVSAGNVRR273 pKa = 11.84QPAMNAKK280 pKa = 7.64MTQKK284 pKa = 10.56CKK286 pKa = 10.82LEE288 pKa = 4.57SGTQLPPRR296 pKa = 11.84TSNEE300 pKa = 3.8AEE302 pKa = 4.09SDD304 pKa = 3.81SEE306 pKa = 4.24YY307 pKa = 11.08DD308 pKa = 4.06DD309 pKa = 4.78EE310 pKa = 5.56LFSEE314 pKa = 4.24IQEE317 pKa = 3.84IRR319 pKa = 11.84SAITKK324 pKa = 8.37LTEE327 pKa = 4.38DD328 pKa = 3.47NQAILTKK335 pKa = 10.44LDD337 pKa = 3.45TLLLLKK343 pKa = 11.03GEE345 pKa = 4.15TDD347 pKa = 3.76SIKK350 pKa = 10.85KK351 pKa = 10.14QISKK355 pKa = 10.63QNIAISTIEE364 pKa = 3.89GHH366 pKa = 6.69LSSIMIAIPGFGKK379 pKa = 9.05DD380 pKa = 3.53TGDD383 pKa = 3.17PTANVDD389 pKa = 3.43INPEE393 pKa = 3.82LRR395 pKa = 11.84PIIGRR400 pKa = 11.84DD401 pKa = 3.02SGRR404 pKa = 11.84ALAEE408 pKa = 4.04VLKK411 pKa = 10.82QPASSRR417 pKa = 11.84GNRR420 pKa = 11.84KK421 pKa = 10.0DD422 pKa = 3.24SGITLGSKK430 pKa = 10.24GQLLRR435 pKa = 11.84DD436 pKa = 3.91LQLKK440 pKa = 9.99PIDD443 pKa = 4.57KK444 pKa = 9.63EE445 pKa = 4.16SSSAIGYY452 pKa = 8.67KK453 pKa = 10.4PKK455 pKa = 9.89DD456 pKa = 3.54TAPSKK461 pKa = 10.96AVLASLIRR469 pKa = 11.84SSRR472 pKa = 11.84VDD474 pKa = 3.13QSHH477 pKa = 5.66KK478 pKa = 10.8HH479 pKa = 5.55NMLALLKK486 pKa = 10.42NIKK489 pKa = 10.53GDD491 pKa = 3.77DD492 pKa = 3.97NLNEE496 pKa = 4.73FYY498 pKa = 11.4QMVKK502 pKa = 10.48SITHH506 pKa = 6.25AA507 pKa = 3.6
MM1 pKa = 8.2AEE3 pKa = 3.69EE4 pKa = 3.84QAYY7 pKa = 9.61HH8 pKa = 5.38VSKK11 pKa = 10.9GLEE14 pKa = 3.92CLKK17 pKa = 10.59ALRR20 pKa = 11.84EE21 pKa = 4.01NPPDD25 pKa = 3.3IEE27 pKa = 4.62EE28 pKa = 4.1IQEE31 pKa = 4.01VSSLRR36 pKa = 11.84DD37 pKa = 3.28QTCNPGQEE45 pKa = 4.05NGTTGMQEE53 pKa = 4.16EE54 pKa = 4.73EE55 pKa = 4.27DD56 pKa = 3.95SQNLDD61 pKa = 3.42EE62 pKa = 4.54SHH64 pKa = 6.74EE65 pKa = 4.09PTKK68 pKa = 10.71GSNYY72 pKa = 9.5VGHH75 pKa = 6.68VPQNNPGCGEE85 pKa = 4.14RR86 pKa = 11.84NTALVEE92 pKa = 4.02AEE94 pKa = 4.06RR95 pKa = 11.84PPRR98 pKa = 11.84EE99 pKa = 4.69DD100 pKa = 3.04IQPGPGIRR108 pKa = 11.84CDD110 pKa = 3.25HH111 pKa = 6.72VYY113 pKa = 10.8DD114 pKa = 4.13HH115 pKa = 7.06SGEE118 pKa = 4.14EE119 pKa = 3.94VKK121 pKa = 11.05GIEE124 pKa = 4.43DD125 pKa = 3.86ADD127 pKa = 4.13SLVVPAGTVGNRR139 pKa = 11.84GFEE142 pKa = 4.04RR143 pKa = 11.84GEE145 pKa = 4.06GSLDD149 pKa = 4.26DD150 pKa = 3.79STEE153 pKa = 4.11DD154 pKa = 3.24SGEE157 pKa = 4.26DD158 pKa = 3.59YY159 pKa = 11.42SEE161 pKa = 4.91GNASSNWGYY170 pKa = 11.52SFGLKK175 pKa = 9.29PDD177 pKa = 3.53RR178 pKa = 11.84AADD181 pKa = 3.34VSMLMEE187 pKa = 4.64EE188 pKa = 4.45EE189 pKa = 4.38LSALLRR195 pKa = 11.84TSRR198 pKa = 11.84NVGIQKK204 pKa = 10.0RR205 pKa = 11.84DD206 pKa = 3.17GKK208 pKa = 8.63TLQFPHH214 pKa = 6.76NPEE217 pKa = 3.84GKK219 pKa = 8.16TRR221 pKa = 11.84DD222 pKa = 4.04PEE224 pKa = 4.44CGSIKK229 pKa = 10.48KK230 pKa = 8.55GTEE233 pKa = 3.79EE234 pKa = 4.29RR235 pKa = 11.84SVSHH239 pKa = 6.08GMGIVAGSTSGATQSALKK257 pKa = 9.2STGGSSEE264 pKa = 4.18PSVSAGNVRR273 pKa = 11.84QPAMNAKK280 pKa = 7.64MTQKK284 pKa = 10.56CKK286 pKa = 10.82LEE288 pKa = 4.57SGTQLPPRR296 pKa = 11.84TSNEE300 pKa = 3.8AEE302 pKa = 4.09SDD304 pKa = 3.81SEE306 pKa = 4.24YY307 pKa = 11.08DD308 pKa = 4.06DD309 pKa = 4.78EE310 pKa = 5.56LFSEE314 pKa = 4.24IQEE317 pKa = 3.84IRR319 pKa = 11.84SAITKK324 pKa = 8.37LTEE327 pKa = 4.38DD328 pKa = 3.47NQAILTKK335 pKa = 10.44LDD337 pKa = 3.45TLLLLKK343 pKa = 11.03GEE345 pKa = 4.15TDD347 pKa = 3.76SIKK350 pKa = 10.85KK351 pKa = 10.14QISKK355 pKa = 10.63QNIAISTIEE364 pKa = 3.89GHH366 pKa = 6.69LSSIMIAIPGFGKK379 pKa = 9.05DD380 pKa = 3.53TGDD383 pKa = 3.17PTANVDD389 pKa = 3.43INPEE393 pKa = 3.82LRR395 pKa = 11.84PIIGRR400 pKa = 11.84DD401 pKa = 3.02SGRR404 pKa = 11.84ALAEE408 pKa = 4.04VLKK411 pKa = 10.82QPASSRR417 pKa = 11.84GNRR420 pKa = 11.84KK421 pKa = 10.0DD422 pKa = 3.24SGITLGSKK430 pKa = 10.24GQLLRR435 pKa = 11.84DD436 pKa = 3.91LQLKK440 pKa = 9.99PIDD443 pKa = 4.57KK444 pKa = 9.63EE445 pKa = 4.16SSSAIGYY452 pKa = 8.67KK453 pKa = 10.4PKK455 pKa = 9.89DD456 pKa = 3.54TAPSKK461 pKa = 10.96AVLASLIRR469 pKa = 11.84SSRR472 pKa = 11.84VDD474 pKa = 3.13QSHH477 pKa = 5.66KK478 pKa = 10.8HH479 pKa = 5.55NMLALLKK486 pKa = 10.42NIKK489 pKa = 10.53GDD491 pKa = 3.77DD492 pKa = 3.97NLNEE496 pKa = 4.73FYY498 pKa = 11.4QMVKK502 pKa = 10.48SITHH506 pKa = 6.25AA507 pKa = 3.6
Molecular weight: 54.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q77Z86|Q77Z86_CDVO Protein C OS=Canine distemper virus (strain Onderstepoort) OX=11233 PE=3 SV=1
MM1 pKa = 7.5SAKK4 pKa = 9.56GWNASKK10 pKa = 10.3PSEE13 pKa = 4.61RR14 pKa = 11.84ILLTLRR20 pKa = 11.84RR21 pKa = 11.84FKK23 pKa = 10.8RR24 pKa = 11.84SAASEE29 pKa = 4.33TKK31 pKa = 10.06PATQAKK37 pKa = 9.67RR38 pKa = 11.84MEE40 pKa = 4.26PQACRR45 pKa = 11.84KK46 pKa = 9.59RR47 pKa = 11.84RR48 pKa = 11.84TLRR51 pKa = 11.84ISMNHH56 pKa = 5.75TSQQKK61 pKa = 9.31DD62 pKa = 3.24QTMSAMYY69 pKa = 9.99LKK71 pKa = 10.34IIRR74 pKa = 11.84DD75 pKa = 3.57VEE77 pKa = 4.08NAILRR82 pKa = 11.84LWRR85 pKa = 11.84RR86 pKa = 11.84SGPLEE91 pKa = 3.8RR92 pKa = 11.84TSNQDD97 pKa = 2.93LEE99 pKa = 4.58YY100 pKa = 10.89DD101 pKa = 3.79VIMFMITAVKK111 pKa = 10.1RR112 pKa = 11.84LRR114 pKa = 11.84EE115 pKa = 4.26SKK117 pKa = 9.8MLTVSWYY124 pKa = 9.9LQALSVIEE132 pKa = 4.25DD133 pKa = 3.33SRR135 pKa = 11.84EE136 pKa = 3.75EE137 pKa = 4.15KK138 pKa = 9.81EE139 pKa = 4.17ALMIALRR146 pKa = 11.84ILAKK150 pKa = 10.28IIPKK154 pKa = 10.22EE155 pKa = 3.92MLHH158 pKa = 5.65LTGDD162 pKa = 3.34ILSALNRR169 pKa = 11.84TEE171 pKa = 3.78QLMM174 pKa = 3.81
MM1 pKa = 7.5SAKK4 pKa = 9.56GWNASKK10 pKa = 10.3PSEE13 pKa = 4.61RR14 pKa = 11.84ILLTLRR20 pKa = 11.84RR21 pKa = 11.84FKK23 pKa = 10.8RR24 pKa = 11.84SAASEE29 pKa = 4.33TKK31 pKa = 10.06PATQAKK37 pKa = 9.67RR38 pKa = 11.84MEE40 pKa = 4.26PQACRR45 pKa = 11.84KK46 pKa = 9.59RR47 pKa = 11.84RR48 pKa = 11.84TLRR51 pKa = 11.84ISMNHH56 pKa = 5.75TSQQKK61 pKa = 9.31DD62 pKa = 3.24QTMSAMYY69 pKa = 9.99LKK71 pKa = 10.34IIRR74 pKa = 11.84DD75 pKa = 3.57VEE77 pKa = 4.08NAILRR82 pKa = 11.84LWRR85 pKa = 11.84RR86 pKa = 11.84SGPLEE91 pKa = 3.8RR92 pKa = 11.84TSNQDD97 pKa = 2.93LEE99 pKa = 4.58YY100 pKa = 10.89DD101 pKa = 3.79VIMFMITAVKK111 pKa = 10.1RR112 pKa = 11.84LRR114 pKa = 11.84EE115 pKa = 4.26SKK117 pKa = 9.8MLTVSWYY124 pKa = 9.9LQALSVIEE132 pKa = 4.25DD133 pKa = 3.33SRR135 pKa = 11.84EE136 pKa = 3.75EE137 pKa = 4.15KK138 pKa = 9.81EE139 pKa = 4.17ALMIALRR146 pKa = 11.84ILAKK150 pKa = 10.28IIPKK154 pKa = 10.22EE155 pKa = 3.92MLHH158 pKa = 5.65LTGDD162 pKa = 3.34ILSALNRR169 pKa = 11.84TEE171 pKa = 3.78QLMM174 pKa = 3.81
Molecular weight: 20.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4989 |
174 |
2184 |
712.7 |
80.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.612 ± 0.374 | 1.744 ± 0.215 |
5.272 ± 0.332 | 5.652 ± 0.526 |
3.387 ± 0.539 | 6.254 ± 0.446 |
2.485 ± 0.392 | 8.138 ± 0.347 |
5.232 ± 0.229 | 10.563 ± 0.384 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.445 ± 0.234 | 4.37 ± 0.102 |
4.39 ± 0.275 | 3.848 ± 0.431 |
5.993 ± 0.245 | 8.88 ± 0.458 |
5.833 ± 0.261 | 5.492 ± 0.295 |
1.002 ± 0.143 | 3.407 ± 0.417 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
