
Capybara microvirus Cap1_SP_22
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
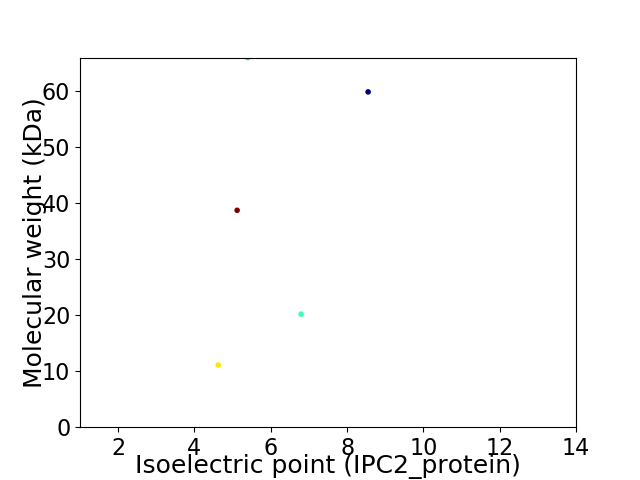
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
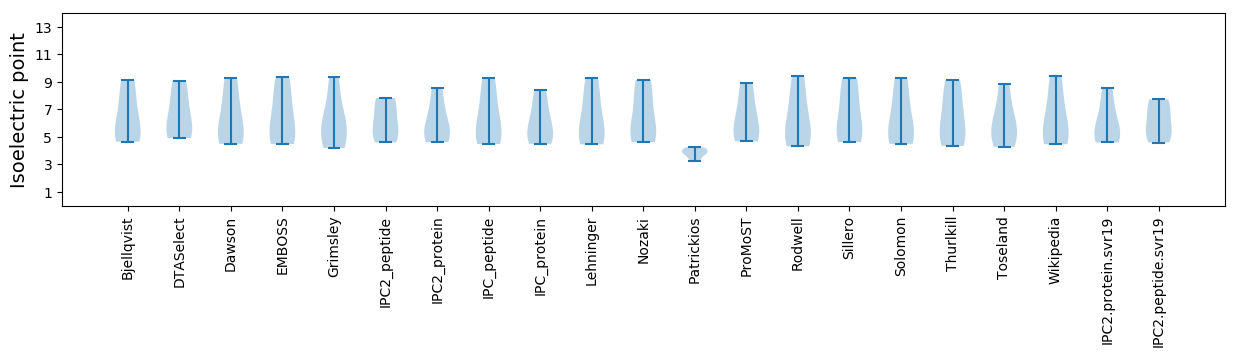
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W4U5|A0A4P8W4U5_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_22 OX=2584777 PE=4 SV=1
MM1 pKa = 7.43TNKK4 pKa = 9.88QKK6 pKa = 10.95FNGVNSFVVKK16 pKa = 10.87LSDD19 pKa = 3.59VPPFSSPTMLYY30 pKa = 10.79CLDD33 pKa = 4.87DD34 pKa = 4.55DD35 pKa = 4.2NKK37 pKa = 10.24VRR39 pKa = 11.84LKK41 pKa = 11.12VVDD44 pKa = 3.18KK45 pKa = 10.57CLNLGPVSNYY55 pKa = 10.63DD56 pKa = 3.65LQTLIDD62 pKa = 3.75NGVNISSMHH71 pKa = 5.99YY72 pKa = 9.99EE73 pKa = 4.15PNDD76 pKa = 3.39DD77 pKa = 3.62RR78 pKa = 11.84LLGVEE83 pKa = 4.63RR84 pKa = 11.84LNTFNPPADD93 pKa = 3.69NDD95 pKa = 4.43SNPNN99 pKa = 3.34
MM1 pKa = 7.43TNKK4 pKa = 9.88QKK6 pKa = 10.95FNGVNSFVVKK16 pKa = 10.87LSDD19 pKa = 3.59VPPFSSPTMLYY30 pKa = 10.79CLDD33 pKa = 4.87DD34 pKa = 4.55DD35 pKa = 4.2NKK37 pKa = 10.24VRR39 pKa = 11.84LKK41 pKa = 11.12VVDD44 pKa = 3.18KK45 pKa = 10.57CLNLGPVSNYY55 pKa = 10.63DD56 pKa = 3.65LQTLIDD62 pKa = 3.75NGVNISSMHH71 pKa = 5.99YY72 pKa = 9.99EE73 pKa = 4.15PNDD76 pKa = 3.39DD77 pKa = 3.62RR78 pKa = 11.84LLGVEE83 pKa = 4.63RR84 pKa = 11.84LNTFNPPADD93 pKa = 3.69NDD95 pKa = 4.43SNPNN99 pKa = 3.34
Molecular weight: 11.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7F3|A0A4P8W7F3_9VIRU Peptidase M15 OS=Capybara microvirus Cap1_SP_22 OX=2584777 PE=4 SV=1
MM1 pKa = 7.31LVSNPTNYY9 pKa = 7.45VTPWQPVMLDD19 pKa = 3.48VPNVRR24 pKa = 11.84SFTYY28 pKa = 10.43LSSLQRR34 pKa = 11.84SYY36 pKa = 11.5YY37 pKa = 8.68IRR39 pKa = 11.84SYY41 pKa = 11.77YY42 pKa = 10.52EE43 pKa = 3.61FLEE46 pKa = 4.4TKK48 pKa = 9.85NRR50 pKa = 11.84NGKK53 pKa = 8.17TIFLTFNYY61 pKa = 9.53NDD63 pKa = 3.34NAIPSIYY70 pKa = 10.44GIPCCNNDD78 pKa = 3.97HH79 pKa = 6.38IHH81 pKa = 5.82YY82 pKa = 10.24LLRR85 pKa = 11.84NSSFIKK91 pKa = 9.73WVRR94 pKa = 11.84QHH96 pKa = 6.83FDD98 pKa = 2.8FSYY101 pKa = 10.4IVCTEE106 pKa = 3.94FGEE109 pKa = 4.85GKK111 pKa = 10.31GKK113 pKa = 10.23RR114 pKa = 11.84GKK116 pKa = 10.62GNNPHH121 pKa = 5.37YY122 pKa = 10.45HH123 pKa = 6.26ALFYY127 pKa = 10.95LCHH130 pKa = 6.98KK131 pKa = 10.26DD132 pKa = 3.48NSQVTPLDD140 pKa = 3.67CAEE143 pKa = 3.95VLSYY147 pKa = 10.23IRR149 pKa = 11.84KK150 pKa = 8.19IWQGNPYY157 pKa = 9.38KK158 pKa = 10.6ASSKK162 pKa = 10.21NDD164 pKa = 2.94PRR166 pKa = 11.84KK167 pKa = 9.78FKK169 pKa = 10.49FGKK172 pKa = 10.19VEE174 pKa = 3.94EE175 pKa = 4.52SEE177 pKa = 4.22KK178 pKa = 11.14GLIVNDD184 pKa = 3.83FHH186 pKa = 7.36AANYY190 pKa = 7.82VSKK193 pKa = 10.94YY194 pKa = 7.19ATKK197 pKa = 10.6DD198 pKa = 3.25KK199 pKa = 8.97EE200 pKa = 4.66TKK202 pKa = 10.42CIIRR206 pKa = 11.84KK207 pKa = 8.87ILNAHH212 pKa = 6.32YY213 pKa = 10.21NDD215 pKa = 4.41CLHH218 pKa = 6.75KK219 pKa = 10.94VEE221 pKa = 4.96LDD223 pKa = 3.18EE224 pKa = 5.7SINHH228 pKa = 5.79TRR230 pKa = 11.84LSLVDD235 pKa = 3.8PFVMPGTTFNDD246 pKa = 3.55KK247 pKa = 10.54VDD249 pKa = 3.36IMVKK253 pKa = 9.85QDD255 pKa = 3.17AQILYY260 pKa = 10.53DD261 pKa = 3.77EE262 pKa = 4.71TLPTIKK268 pKa = 9.52KK269 pKa = 7.71TFLPKK274 pKa = 10.53VFMSKK279 pKa = 10.41GFGSSASKK287 pKa = 11.03YY288 pKa = 8.71ITDD291 pKa = 4.06DD292 pKa = 3.24FKK294 pKa = 11.46MPILTHH300 pKa = 6.61KK301 pKa = 8.96GTIYY305 pKa = 9.7VTLPYY310 pKa = 10.17YY311 pKa = 10.21IYY313 pKa = 10.73RR314 pKa = 11.84KK315 pKa = 7.8TFYY318 pKa = 10.42DD319 pKa = 3.38VKK321 pKa = 10.96KK322 pKa = 9.92NANGNVIYY330 pKa = 10.6VLNNVGTEE338 pKa = 3.85YY339 pKa = 10.79RR340 pKa = 11.84KK341 pKa = 10.04NRR343 pKa = 11.84LSKK346 pKa = 10.45DD347 pKa = 2.96IRR349 pKa = 11.84DD350 pKa = 4.29LYY352 pKa = 10.68SKK354 pKa = 10.49CISILHH360 pKa = 6.24NSDD363 pKa = 3.2TLSNNDD369 pKa = 3.58SEE371 pKa = 4.38EE372 pKa = 4.46DD373 pKa = 3.13IYY375 pKa = 11.76ANKK378 pKa = 9.36IRR380 pKa = 11.84HH381 pKa = 5.53LHH383 pKa = 6.26HH384 pKa = 6.51SLKK387 pKa = 10.5SHH389 pKa = 6.07VSAPFLDD396 pKa = 4.25PMHH399 pKa = 7.22AYY401 pKa = 10.26ASYY404 pKa = 10.66KK405 pKa = 10.26YY406 pKa = 9.45IYY408 pKa = 9.78QYY410 pKa = 11.27RR411 pKa = 11.84CFEE414 pKa = 4.1GLEE417 pKa = 4.23FPLDD421 pKa = 4.03LNDD424 pKa = 5.31DD425 pKa = 4.09FALKK429 pKa = 10.43LLPGYY434 pKa = 10.73YY435 pKa = 10.0YY436 pKa = 11.35GEE438 pKa = 4.37FSNSPILTYY447 pKa = 10.65KK448 pKa = 10.61EE449 pKa = 3.89LTKK452 pKa = 11.07SNWHH456 pKa = 6.72PFSEE460 pKa = 4.47HH461 pKa = 6.37SSFVPLIPLFNVLDD475 pKa = 3.96KK476 pKa = 11.64YY477 pKa = 11.24FDD479 pKa = 3.74LVGIQLDD486 pKa = 3.44DD487 pKa = 3.63ARR489 pKa = 11.84YY490 pKa = 9.85KK491 pKa = 10.51RR492 pKa = 11.84YY493 pKa = 9.57LRR495 pKa = 11.84KK496 pKa = 10.11KK497 pKa = 8.22MLFNFFSKK505 pKa = 10.69YY506 pKa = 8.9KK507 pKa = 10.44RR508 pKa = 11.84KK509 pKa = 10.1
MM1 pKa = 7.31LVSNPTNYY9 pKa = 7.45VTPWQPVMLDD19 pKa = 3.48VPNVRR24 pKa = 11.84SFTYY28 pKa = 10.43LSSLQRR34 pKa = 11.84SYY36 pKa = 11.5YY37 pKa = 8.68IRR39 pKa = 11.84SYY41 pKa = 11.77YY42 pKa = 10.52EE43 pKa = 3.61FLEE46 pKa = 4.4TKK48 pKa = 9.85NRR50 pKa = 11.84NGKK53 pKa = 8.17TIFLTFNYY61 pKa = 9.53NDD63 pKa = 3.34NAIPSIYY70 pKa = 10.44GIPCCNNDD78 pKa = 3.97HH79 pKa = 6.38IHH81 pKa = 5.82YY82 pKa = 10.24LLRR85 pKa = 11.84NSSFIKK91 pKa = 9.73WVRR94 pKa = 11.84QHH96 pKa = 6.83FDD98 pKa = 2.8FSYY101 pKa = 10.4IVCTEE106 pKa = 3.94FGEE109 pKa = 4.85GKK111 pKa = 10.31GKK113 pKa = 10.23RR114 pKa = 11.84GKK116 pKa = 10.62GNNPHH121 pKa = 5.37YY122 pKa = 10.45HH123 pKa = 6.26ALFYY127 pKa = 10.95LCHH130 pKa = 6.98KK131 pKa = 10.26DD132 pKa = 3.48NSQVTPLDD140 pKa = 3.67CAEE143 pKa = 3.95VLSYY147 pKa = 10.23IRR149 pKa = 11.84KK150 pKa = 8.19IWQGNPYY157 pKa = 9.38KK158 pKa = 10.6ASSKK162 pKa = 10.21NDD164 pKa = 2.94PRR166 pKa = 11.84KK167 pKa = 9.78FKK169 pKa = 10.49FGKK172 pKa = 10.19VEE174 pKa = 3.94EE175 pKa = 4.52SEE177 pKa = 4.22KK178 pKa = 11.14GLIVNDD184 pKa = 3.83FHH186 pKa = 7.36AANYY190 pKa = 7.82VSKK193 pKa = 10.94YY194 pKa = 7.19ATKK197 pKa = 10.6DD198 pKa = 3.25KK199 pKa = 8.97EE200 pKa = 4.66TKK202 pKa = 10.42CIIRR206 pKa = 11.84KK207 pKa = 8.87ILNAHH212 pKa = 6.32YY213 pKa = 10.21NDD215 pKa = 4.41CLHH218 pKa = 6.75KK219 pKa = 10.94VEE221 pKa = 4.96LDD223 pKa = 3.18EE224 pKa = 5.7SINHH228 pKa = 5.79TRR230 pKa = 11.84LSLVDD235 pKa = 3.8PFVMPGTTFNDD246 pKa = 3.55KK247 pKa = 10.54VDD249 pKa = 3.36IMVKK253 pKa = 9.85QDD255 pKa = 3.17AQILYY260 pKa = 10.53DD261 pKa = 3.77EE262 pKa = 4.71TLPTIKK268 pKa = 9.52KK269 pKa = 7.71TFLPKK274 pKa = 10.53VFMSKK279 pKa = 10.41GFGSSASKK287 pKa = 11.03YY288 pKa = 8.71ITDD291 pKa = 4.06DD292 pKa = 3.24FKK294 pKa = 11.46MPILTHH300 pKa = 6.61KK301 pKa = 8.96GTIYY305 pKa = 9.7VTLPYY310 pKa = 10.17YY311 pKa = 10.21IYY313 pKa = 10.73RR314 pKa = 11.84KK315 pKa = 7.8TFYY318 pKa = 10.42DD319 pKa = 3.38VKK321 pKa = 10.96KK322 pKa = 9.92NANGNVIYY330 pKa = 10.6VLNNVGTEE338 pKa = 3.85YY339 pKa = 10.79RR340 pKa = 11.84KK341 pKa = 10.04NRR343 pKa = 11.84LSKK346 pKa = 10.45DD347 pKa = 2.96IRR349 pKa = 11.84DD350 pKa = 4.29LYY352 pKa = 10.68SKK354 pKa = 10.49CISILHH360 pKa = 6.24NSDD363 pKa = 3.2TLSNNDD369 pKa = 3.58SEE371 pKa = 4.38EE372 pKa = 4.46DD373 pKa = 3.13IYY375 pKa = 11.76ANKK378 pKa = 9.36IRR380 pKa = 11.84HH381 pKa = 5.53LHH383 pKa = 6.26HH384 pKa = 6.51SLKK387 pKa = 10.5SHH389 pKa = 6.07VSAPFLDD396 pKa = 4.25PMHH399 pKa = 7.22AYY401 pKa = 10.26ASYY404 pKa = 10.66KK405 pKa = 10.26YY406 pKa = 9.45IYY408 pKa = 9.78QYY410 pKa = 11.27RR411 pKa = 11.84CFEE414 pKa = 4.1GLEE417 pKa = 4.23FPLDD421 pKa = 4.03LNDD424 pKa = 5.31DD425 pKa = 4.09FALKK429 pKa = 10.43LLPGYY434 pKa = 10.73YY435 pKa = 10.0YY436 pKa = 11.35GEE438 pKa = 4.37FSNSPILTYY447 pKa = 10.65KK448 pKa = 10.61EE449 pKa = 3.89LTKK452 pKa = 11.07SNWHH456 pKa = 6.72PFSEE460 pKa = 4.47HH461 pKa = 6.37SSFVPLIPLFNVLDD475 pKa = 3.96KK476 pKa = 11.64YY477 pKa = 11.24FDD479 pKa = 3.74LVGIQLDD486 pKa = 3.44DD487 pKa = 3.63ARR489 pKa = 11.84YY490 pKa = 9.85KK491 pKa = 10.51RR492 pKa = 11.84YY493 pKa = 9.57LRR495 pKa = 11.84KK496 pKa = 10.11KK497 pKa = 8.22MLFNFFSKK505 pKa = 10.69YY506 pKa = 8.9KK507 pKa = 10.44RR508 pKa = 11.84KK509 pKa = 10.1
Molecular weight: 59.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1713 |
99 |
583 |
342.6 |
39.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.546 ± 1.268 | 1.693 ± 0.428 |
6.421 ± 0.612 | 4.32 ± 0.789 |
5.838 ± 0.889 | 5.137 ± 0.605 |
2.393 ± 0.568 | 5.721 ± 0.397 |
6.83 ± 0.874 | 9.048 ± 0.302 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.926 ± 0.353 | 7.823 ± 0.883 |
3.853 ± 0.614 | 3.736 ± 1.085 |
3.678 ± 0.256 | 8.173 ± 0.624 |
6.305 ± 0.698 | 4.845 ± 0.737 |
0.992 ± 0.304 | 5.721 ± 1.095 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
