
Cyclobacterium marinum (strain ATCC 25205 / DSM 745 / LMG 13164 / NCIMB 1802) (Flectobacillus marinus)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Cyclobacteriaceae; Cyclobacterium; Cyclobacterium marinum
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
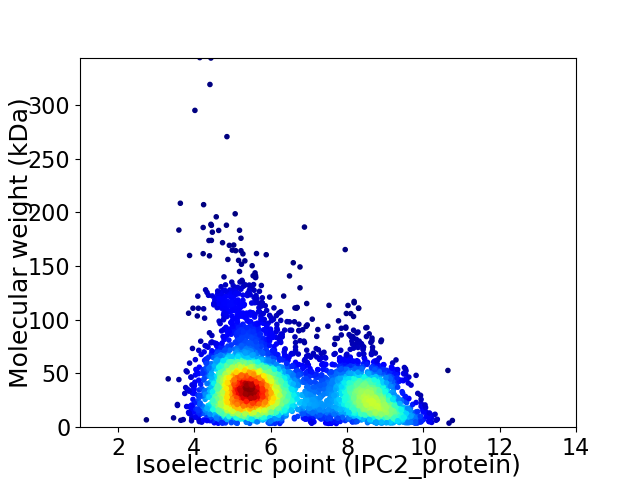
Virtual 2D-PAGE plot for 4983 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G0J4Y2|G0J4Y2_CYCMS Uncharacterized protein OS=Cyclobacterium marinum (strain ATCC 25205 / DSM 745 / LMG 13164 / NCIMB 1802) OX=880070 GN=Cycma_2234 PE=4 SV=1
MM1 pKa = 7.54KK2 pKa = 10.31SFHH5 pKa = 6.71KK6 pKa = 10.19FVLVIVSISFFQTGCNTGMDD26 pKa = 3.95EE27 pKa = 5.43DD28 pKa = 4.33IPSISDD34 pKa = 3.47YY35 pKa = 11.34SGTASISQGFATATINNLFEE55 pKa = 4.54CSNGRR60 pKa = 11.84DD61 pKa = 3.59APLGISTAMDD71 pKa = 4.22GSTWTVPAQVNFNDD85 pKa = 3.85SNFPFASDD93 pKa = 4.68LYY95 pKa = 9.79NPCSGLTFNTDD106 pKa = 3.07TEE108 pKa = 4.35AVLALKK114 pKa = 10.67DD115 pKa = 3.59EE116 pKa = 5.53DD117 pKa = 4.45IIEE120 pKa = 4.14VDD122 pKa = 3.76PNGEE126 pKa = 4.51LITAFVFADD135 pKa = 4.12NYY137 pKa = 10.72FEE139 pKa = 4.36MYY141 pKa = 10.37INGVPVGKK149 pKa = 10.35DD150 pKa = 3.04MVPFTPFNSSILRR163 pKa = 11.84FRR165 pKa = 11.84VSYY168 pKa = 10.38PFTIAMKK175 pKa = 10.59LVDD178 pKa = 3.52WEE180 pKa = 4.48EE181 pKa = 3.85NSGLGSEE188 pKa = 4.62SNQGASFYY196 pKa = 10.38PGDD199 pKa = 3.78GGMVAVFKK207 pKa = 10.9NQANEE212 pKa = 3.83IVATTGADD220 pKa = 2.85WKK222 pKa = 10.91AQTFYY227 pKa = 10.9TSPIKK232 pKa = 10.51DD233 pKa = 3.9LSCTTEE239 pKa = 3.61NGTQRR244 pKa = 11.84LSYY247 pKa = 10.73NCNTDD252 pKa = 2.77GANDD256 pKa = 3.87GTSFYY261 pKa = 10.92ALHH264 pKa = 6.85WDD266 pKa = 4.46IPANWQEE273 pKa = 3.81EE274 pKa = 4.28DD275 pKa = 4.17FDD277 pKa = 5.16DD278 pKa = 5.46SSWPNATPYY287 pKa = 10.38TNTEE291 pKa = 3.36IGVNNKK297 pKa = 8.81PAYY300 pKa = 9.55TNFTSIFDD308 pKa = 4.18DD309 pKa = 5.32PIDD312 pKa = 4.0DD313 pKa = 5.89AEE315 pKa = 5.13FIWSTNVILDD325 pKa = 3.38NEE327 pKa = 4.63VIVRR331 pKa = 11.84FTVEE335 pKa = 3.35
MM1 pKa = 7.54KK2 pKa = 10.31SFHH5 pKa = 6.71KK6 pKa = 10.19FVLVIVSISFFQTGCNTGMDD26 pKa = 3.95EE27 pKa = 5.43DD28 pKa = 4.33IPSISDD34 pKa = 3.47YY35 pKa = 11.34SGTASISQGFATATINNLFEE55 pKa = 4.54CSNGRR60 pKa = 11.84DD61 pKa = 3.59APLGISTAMDD71 pKa = 4.22GSTWTVPAQVNFNDD85 pKa = 3.85SNFPFASDD93 pKa = 4.68LYY95 pKa = 9.79NPCSGLTFNTDD106 pKa = 3.07TEE108 pKa = 4.35AVLALKK114 pKa = 10.67DD115 pKa = 3.59EE116 pKa = 5.53DD117 pKa = 4.45IIEE120 pKa = 4.14VDD122 pKa = 3.76PNGEE126 pKa = 4.51LITAFVFADD135 pKa = 4.12NYY137 pKa = 10.72FEE139 pKa = 4.36MYY141 pKa = 10.37INGVPVGKK149 pKa = 10.35DD150 pKa = 3.04MVPFTPFNSSILRR163 pKa = 11.84FRR165 pKa = 11.84VSYY168 pKa = 10.38PFTIAMKK175 pKa = 10.59LVDD178 pKa = 3.52WEE180 pKa = 4.48EE181 pKa = 3.85NSGLGSEE188 pKa = 4.62SNQGASFYY196 pKa = 10.38PGDD199 pKa = 3.78GGMVAVFKK207 pKa = 10.9NQANEE212 pKa = 3.83IVATTGADD220 pKa = 2.85WKK222 pKa = 10.91AQTFYY227 pKa = 10.9TSPIKK232 pKa = 10.51DD233 pKa = 3.9LSCTTEE239 pKa = 3.61NGTQRR244 pKa = 11.84LSYY247 pKa = 10.73NCNTDD252 pKa = 2.77GANDD256 pKa = 3.87GTSFYY261 pKa = 10.92ALHH264 pKa = 6.85WDD266 pKa = 4.46IPANWQEE273 pKa = 3.81EE274 pKa = 4.28DD275 pKa = 4.17FDD277 pKa = 5.16DD278 pKa = 5.46SSWPNATPYY287 pKa = 10.38TNTEE291 pKa = 3.36IGVNNKK297 pKa = 8.81PAYY300 pKa = 9.55TNFTSIFDD308 pKa = 4.18DD309 pKa = 5.32PIDD312 pKa = 4.0DD313 pKa = 5.89AEE315 pKa = 5.13FIWSTNVILDD325 pKa = 3.38NEE327 pKa = 4.63VIVRR331 pKa = 11.84FTVEE335 pKa = 3.35
Molecular weight: 36.91 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G0IYF5|G0IYF5_CYCMS Transcription termination/antitermination protein NusA OS=Cyclobacterium marinum (strain ATCC 25205 / DSM 745 / LMG 13164 / NCIMB 1802) OX=880070 GN=nusA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.5HH16 pKa = 3.99GFRR19 pKa = 11.84EE20 pKa = 4.15RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.5ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.17GRR39 pKa = 11.84HH40 pKa = 5.24KK41 pKa = 10.87LSVSSEE47 pKa = 3.97KK48 pKa = 9.91TLKK51 pKa = 10.51KK52 pKa = 10.72
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.62RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.5HH16 pKa = 3.99GFRR19 pKa = 11.84EE20 pKa = 4.15RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84RR29 pKa = 11.84VLKK32 pKa = 10.5ARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.17GRR39 pKa = 11.84HH40 pKa = 5.24KK41 pKa = 10.87LSVSSEE47 pKa = 3.97KK48 pKa = 9.91TLKK51 pKa = 10.51KK52 pKa = 10.72
Molecular weight: 6.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1776705 |
30 |
3154 |
356.6 |
40.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.469 ± 0.034 | 0.695 ± 0.011 |
5.443 ± 0.022 | 6.869 ± 0.027 |
5.137 ± 0.024 | 7.036 ± 0.034 |
1.846 ± 0.015 | 7.397 ± 0.031 |
7.117 ± 0.038 | 9.732 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.417 ± 0.018 | 5.67 ± 0.035 |
3.945 ± 0.021 | 3.504 ± 0.018 |
3.763 ± 0.02 | 6.594 ± 0.025 |
5.075 ± 0.021 | 6.053 ± 0.024 |
1.303 ± 0.015 | 3.935 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
