
Ruegeria sp. (strain TM1040) (Silicibacter sp.)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Ruegeria; unclassified Ruegeria
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
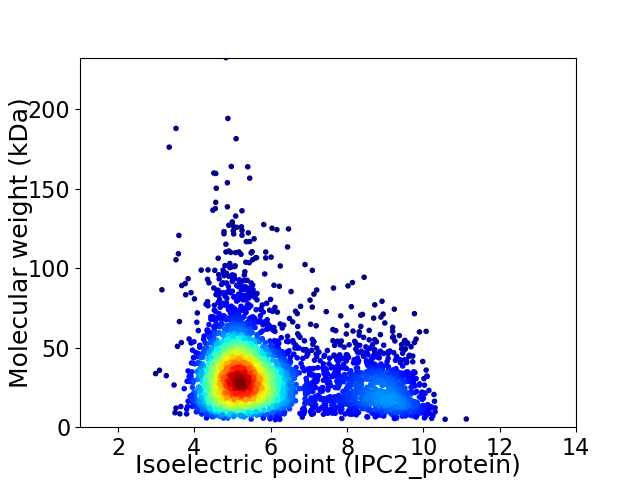
Virtual 2D-PAGE plot for 3849 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q1GM70|Q1GM70_RUEST Thiolase OS=Ruegeria sp. (strain TM1040) OX=292414 GN=TM1040_3272 PE=3 SV=1
MM1 pKa = 8.28IEE3 pKa = 4.28FLCEE7 pKa = 3.55CVVRR11 pKa = 11.84DD12 pKa = 4.44KK13 pKa = 10.92RR14 pKa = 11.84CAMTPPVSGYY24 pKa = 8.28KK25 pKa = 9.2TGRR28 pKa = 11.84GPIVKK33 pKa = 9.6RR34 pKa = 11.84LSGVFFFIMVSLFVAPQAAQASFTSCPANTSGGDD68 pKa = 3.82PLTMNLTVDD77 pKa = 3.45EE78 pKa = 4.78CTLQSGAAVSALSDD92 pKa = 4.1DD93 pKa = 5.51LIQIQIAGGSSTNFFIFDD111 pKa = 4.14PSANDD116 pKa = 3.31LRR118 pKa = 11.84DD119 pKa = 4.04LIFSVNNGSPQGTLDD134 pKa = 3.7ADD136 pKa = 3.94GGSLSGIDD144 pKa = 3.67CAAGCTVSGTHH155 pKa = 6.28GGSPFTFTYY164 pKa = 8.54TASGGGGSLSSPAEE178 pKa = 3.74IDD180 pKa = 3.95DD181 pKa = 4.39LVSSTGNTVADD192 pKa = 4.31GGTDD196 pKa = 4.05TIAAAIDD203 pKa = 3.6SGTPTTITYY212 pKa = 9.97DD213 pKa = 3.45VSNSGSGTLSLSGLTQGTPTNASFGATSIDD243 pKa = 3.61AASVASGGTEE253 pKa = 3.49TDD255 pKa = 3.85AISITFTPSSTTDD268 pKa = 3.16GASFSVPFSFTTNDD282 pKa = 3.39ADD284 pKa = 5.03DD285 pKa = 4.66GTDD288 pKa = 2.94EE289 pKa = 4.31TTFNVTISGTINAEE303 pKa = 3.96AEE305 pKa = 4.18ISDD308 pKa = 4.13LTSSTTNTVADD319 pKa = 4.6GGSDD323 pKa = 3.41AQGTVASGSAQALIYY338 pKa = 10.11TITNSGTADD347 pKa = 3.63LSVATASSSSASNVTVNSIGAPGSTTVSAGGGTTQFTVQYY387 pKa = 8.78TPSAAGAFSFDD398 pKa = 3.41LSFVNSDD405 pKa = 3.2ADD407 pKa = 3.51EE408 pKa = 4.98DD409 pKa = 4.29PFNFTVSGTATAAPGFSQAIAPGTILADD437 pKa = 3.5GTATVTFTIDD447 pKa = 2.78NSANATAATSLDD459 pKa = 3.75FSNSLPAGVEE469 pKa = 4.17VAATPNASTTCTGGTLTAVAGSGTLSYY496 pKa = 10.71TGGTVGAGASCTVQADD512 pKa = 3.68VTAGTDD518 pKa = 3.41GSYY521 pKa = 11.35SNTSGDD527 pKa = 3.75LTSSLGNSGSASASLTVASPEE548 pKa = 3.83IDD550 pKa = 3.57LQRR553 pKa = 11.84PAATSLADD561 pKa = 4.06GDD563 pKa = 3.9TDD565 pKa = 3.52AQGNVAVGVQQVLTYY580 pKa = 9.89TVEE583 pKa = 4.08NTGNATLTLSGTPSSSAASNVSVDD607 pKa = 4.28SISAPGSSSLAASASTTFTVAYY629 pKa = 8.05TPTAAGAFSFEE640 pKa = 5.13LDD642 pKa = 3.71VSSDD646 pKa = 3.62DD647 pKa = 4.37ADD649 pKa = 3.48EE650 pKa = 4.38GTYY653 pKa = 11.13DD654 pKa = 3.64LTVSGTATAAPGFSQAIAPGTILADD679 pKa = 3.5GTATVTFTIDD689 pKa = 2.78NSANATAATSLDD701 pKa = 3.75FSNSLPAGVEE711 pKa = 4.17VAATPNAATSCTGGTLTAVAGSGTLSYY738 pKa = 10.71TGGTVGAGASCTVQADD754 pKa = 3.68VTAGTDD760 pKa = 3.41GSYY763 pKa = 11.35SNTSGDD769 pKa = 3.75LTSSLGNSGSASASLTVASPEE790 pKa = 3.83IDD792 pKa = 3.57LQRR795 pKa = 11.84PAATSLADD803 pKa = 4.06GDD805 pKa = 3.9TDD807 pKa = 3.52AQGNVAVGVQQVLTYY822 pKa = 9.89TVEE825 pKa = 4.08NTGNATLTLSGTPSSAAASNVSVDD849 pKa = 4.28SISAPGSSSLAASASTTFTVAYY871 pKa = 8.05TPTAAGAFSFEE882 pKa = 5.13LDD884 pKa = 3.71VSSDD888 pKa = 3.62DD889 pKa = 4.37ADD891 pKa = 3.48EE892 pKa = 4.38GTYY895 pKa = 11.13DD896 pKa = 3.64LTVSGTATAAPGFSQAIAPGTILADD921 pKa = 3.5GTATVTFTIDD931 pKa = 2.78NSANATAATSLDD943 pKa = 3.75FSNSLPAGVEE953 pKa = 4.17VAATPNAATSCTGGTLTAVAGSGTLSYY980 pKa = 10.71TGGTVGAGASCTVQADD996 pKa = 3.68VTAGTDD1002 pKa = 3.41GSYY1005 pKa = 11.35SNTSGDD1011 pKa = 3.75LTSSLGNSGSASASLTVASPEE1032 pKa = 3.83IDD1034 pKa = 3.57LQRR1037 pKa = 11.84PAATSLADD1045 pKa = 4.06GDD1047 pKa = 3.9TDD1049 pKa = 3.52AQGNVAVGVQQVLTYY1064 pKa = 9.6TVEE1067 pKa = 4.15NTGTATLTLSGTPSSAAASNVSVDD1091 pKa = 4.28SISAPGSSSLAASASTTFTVAYY1113 pKa = 8.05TPTAAGAFSFEE1124 pKa = 5.13LDD1126 pKa = 3.71VSSDD1130 pKa = 3.62DD1131 pKa = 4.37ADD1133 pKa = 3.48EE1134 pKa = 4.38GTYY1137 pKa = 11.13DD1138 pKa = 3.64LTVSGTADD1146 pKa = 3.34GSAEE1150 pKa = 4.0IGVGSATGGSLEE1162 pKa = 5.79DD1163 pKa = 4.69GDD1165 pKa = 4.71TDD1167 pKa = 4.78TIPGTPSAGSPTVIIYY1183 pKa = 7.5TFTNSGSGTLNITTPTVAGNISNEE1207 pKa = 4.13TNVVVNSLTLASNTVAGGGGTTTLEE1232 pKa = 3.54ISYY1235 pKa = 8.8TPQVSGAYY1243 pKa = 10.05RR1244 pKa = 11.84FDD1246 pKa = 3.66LQVANDD1252 pKa = 4.4DD1253 pKa = 3.9ADD1255 pKa = 3.46EE1256 pKa = 4.33SLFNITVAGSASGLPEE1272 pKa = 4.42IEE1274 pKa = 4.43VSSSASGALSDD1285 pKa = 5.17GDD1287 pKa = 3.73TDD1289 pKa = 5.08SIATVATPGVTASVTYY1305 pKa = 9.93TITNSGTDD1313 pKa = 3.74VLTLDD1318 pKa = 4.26APSVASNIRR1327 pKa = 11.84NQSNVTVDD1335 pKa = 3.68RR1336 pKa = 11.84LSLASTTVASGGGTTTLRR1354 pKa = 11.84VDD1356 pKa = 3.53YY1357 pKa = 8.79TATAGGAYY1365 pKa = 10.11GFDD1368 pKa = 3.94LSLGSDD1374 pKa = 4.38DD1375 pKa = 5.62ADD1377 pKa = 3.5EE1378 pKa = 5.14DD1379 pKa = 3.96PFDD1382 pKa = 3.77IVVSGTAQSVASALAAQSGSGQVSEE1407 pKa = 5.29VGAQFPAPLVVLVTDD1422 pKa = 3.64STGTGVAGVDD1432 pKa = 3.36VTFAAPASGASVVFASTGTHH1452 pKa = 5.82TEE1454 pKa = 4.15TVTTGSDD1461 pKa = 3.31GTASTSAMTANGTPSSYY1478 pKa = 11.34AGGSTLQSYY1487 pKa = 10.87DD1488 pKa = 3.22VTASATGLASVTFALTNDD1506 pKa = 3.81RR1507 pKa = 11.84DD1508 pKa = 3.95SEE1510 pKa = 4.18ADD1512 pKa = 3.08IQKK1515 pKa = 7.76TQEE1518 pKa = 3.77VIAAFVSNRR1527 pKa = 11.84ANAIVSNQPDD1537 pKa = 3.74LVSRR1541 pKa = 11.84LRR1543 pKa = 11.84QGALAGQQGRR1553 pKa = 11.84NGFDD1557 pKa = 3.13LRR1559 pKa = 11.84ATSGGRR1565 pKa = 11.84SANFSFSLRR1574 pKa = 11.84ALLEE1578 pKa = 3.74EE1579 pKa = 3.91ARR1581 pKa = 11.84RR1582 pKa = 11.84RR1583 pKa = 11.84NDD1585 pKa = 3.03GARR1588 pKa = 11.84WRR1590 pKa = 11.84EE1591 pKa = 3.99TLRR1594 pKa = 11.84HH1595 pKa = 6.08ADD1597 pKa = 3.27ASAGPGADD1605 pKa = 3.25SAADD1609 pKa = 3.42RR1610 pKa = 11.84AMASQAYY1617 pKa = 9.3ALRR1620 pKa = 11.84FRR1622 pKa = 11.84GTDD1625 pKa = 3.38PAQAAPVVTPVISSGWDD1642 pKa = 3.39FWAEE1646 pKa = 3.4GTYY1649 pKa = 10.55AITEE1653 pKa = 4.2SGNFEE1658 pKa = 4.15SRR1660 pKa = 11.84SGLFFAGADD1669 pKa = 3.93YY1670 pKa = 10.56RR1671 pKa = 11.84WSDD1674 pKa = 3.63QVLLGVMGQLDD1685 pKa = 3.72IADD1688 pKa = 4.48EE1689 pKa = 4.76DD1690 pKa = 3.89NVAAGTSASGTGWMVGPYY1708 pKa = 9.93VVVRR1712 pKa = 11.84LDD1714 pKa = 3.18QNLYY1718 pKa = 10.34LDD1720 pKa = 3.63VAATYY1725 pKa = 10.14GRR1727 pKa = 11.84SSNTVNALGLFEE1739 pKa = 6.72DD1740 pKa = 5.12DD1741 pKa = 4.4FDD1743 pKa = 4.21TEE1745 pKa = 4.43RR1746 pKa = 11.84FLLQGGLTGDD1756 pKa = 4.25FKK1758 pKa = 11.72LNARR1762 pKa = 11.84TTISPFARR1770 pKa = 11.84LTYY1773 pKa = 10.12YY1774 pKa = 10.31YY1775 pKa = 9.6EE1776 pKa = 4.23KK1777 pKa = 10.59QEE1779 pKa = 4.49SYY1781 pKa = 10.99TDD1783 pKa = 3.27TLGRR1787 pKa = 11.84VIPSQDD1793 pKa = 3.07FEE1795 pKa = 4.64LGRR1798 pKa = 11.84FEE1800 pKa = 4.84FGPKK1804 pKa = 9.75ISWEE1808 pKa = 4.16MYY1810 pKa = 9.38LQEE1813 pKa = 4.36GTQFSPYY1820 pKa = 10.25VSFSGIYY1827 pKa = 10.21DD1828 pKa = 3.59FNKK1831 pKa = 10.39LQGATPTDD1839 pKa = 3.28ATLASSQSDD1848 pKa = 3.24LRR1850 pKa = 11.84GRR1852 pKa = 11.84LEE1854 pKa = 4.12AGATFLVPDD1863 pKa = 4.45RR1864 pKa = 11.84GIKK1867 pKa = 9.53VAVEE1871 pKa = 3.91GFYY1874 pKa = 11.15DD1875 pKa = 5.3GIGTSDD1881 pKa = 3.76FEE1883 pKa = 4.93SYY1885 pKa = 10.6GASLSVLIPFF1895 pKa = 5.19
MM1 pKa = 8.28IEE3 pKa = 4.28FLCEE7 pKa = 3.55CVVRR11 pKa = 11.84DD12 pKa = 4.44KK13 pKa = 10.92RR14 pKa = 11.84CAMTPPVSGYY24 pKa = 8.28KK25 pKa = 9.2TGRR28 pKa = 11.84GPIVKK33 pKa = 9.6RR34 pKa = 11.84LSGVFFFIMVSLFVAPQAAQASFTSCPANTSGGDD68 pKa = 3.82PLTMNLTVDD77 pKa = 3.45EE78 pKa = 4.78CTLQSGAAVSALSDD92 pKa = 4.1DD93 pKa = 5.51LIQIQIAGGSSTNFFIFDD111 pKa = 4.14PSANDD116 pKa = 3.31LRR118 pKa = 11.84DD119 pKa = 4.04LIFSVNNGSPQGTLDD134 pKa = 3.7ADD136 pKa = 3.94GGSLSGIDD144 pKa = 3.67CAAGCTVSGTHH155 pKa = 6.28GGSPFTFTYY164 pKa = 8.54TASGGGGSLSSPAEE178 pKa = 3.74IDD180 pKa = 3.95DD181 pKa = 4.39LVSSTGNTVADD192 pKa = 4.31GGTDD196 pKa = 4.05TIAAAIDD203 pKa = 3.6SGTPTTITYY212 pKa = 9.97DD213 pKa = 3.45VSNSGSGTLSLSGLTQGTPTNASFGATSIDD243 pKa = 3.61AASVASGGTEE253 pKa = 3.49TDD255 pKa = 3.85AISITFTPSSTTDD268 pKa = 3.16GASFSVPFSFTTNDD282 pKa = 3.39ADD284 pKa = 5.03DD285 pKa = 4.66GTDD288 pKa = 2.94EE289 pKa = 4.31TTFNVTISGTINAEE303 pKa = 3.96AEE305 pKa = 4.18ISDD308 pKa = 4.13LTSSTTNTVADD319 pKa = 4.6GGSDD323 pKa = 3.41AQGTVASGSAQALIYY338 pKa = 10.11TITNSGTADD347 pKa = 3.63LSVATASSSSASNVTVNSIGAPGSTTVSAGGGTTQFTVQYY387 pKa = 8.78TPSAAGAFSFDD398 pKa = 3.41LSFVNSDD405 pKa = 3.2ADD407 pKa = 3.51EE408 pKa = 4.98DD409 pKa = 4.29PFNFTVSGTATAAPGFSQAIAPGTILADD437 pKa = 3.5GTATVTFTIDD447 pKa = 2.78NSANATAATSLDD459 pKa = 3.75FSNSLPAGVEE469 pKa = 4.17VAATPNASTTCTGGTLTAVAGSGTLSYY496 pKa = 10.71TGGTVGAGASCTVQADD512 pKa = 3.68VTAGTDD518 pKa = 3.41GSYY521 pKa = 11.35SNTSGDD527 pKa = 3.75LTSSLGNSGSASASLTVASPEE548 pKa = 3.83IDD550 pKa = 3.57LQRR553 pKa = 11.84PAATSLADD561 pKa = 4.06GDD563 pKa = 3.9TDD565 pKa = 3.52AQGNVAVGVQQVLTYY580 pKa = 9.89TVEE583 pKa = 4.08NTGNATLTLSGTPSSSAASNVSVDD607 pKa = 4.28SISAPGSSSLAASASTTFTVAYY629 pKa = 8.05TPTAAGAFSFEE640 pKa = 5.13LDD642 pKa = 3.71VSSDD646 pKa = 3.62DD647 pKa = 4.37ADD649 pKa = 3.48EE650 pKa = 4.38GTYY653 pKa = 11.13DD654 pKa = 3.64LTVSGTATAAPGFSQAIAPGTILADD679 pKa = 3.5GTATVTFTIDD689 pKa = 2.78NSANATAATSLDD701 pKa = 3.75FSNSLPAGVEE711 pKa = 4.17VAATPNAATSCTGGTLTAVAGSGTLSYY738 pKa = 10.71TGGTVGAGASCTVQADD754 pKa = 3.68VTAGTDD760 pKa = 3.41GSYY763 pKa = 11.35SNTSGDD769 pKa = 3.75LTSSLGNSGSASASLTVASPEE790 pKa = 3.83IDD792 pKa = 3.57LQRR795 pKa = 11.84PAATSLADD803 pKa = 4.06GDD805 pKa = 3.9TDD807 pKa = 3.52AQGNVAVGVQQVLTYY822 pKa = 9.89TVEE825 pKa = 4.08NTGNATLTLSGTPSSAAASNVSVDD849 pKa = 4.28SISAPGSSSLAASASTTFTVAYY871 pKa = 8.05TPTAAGAFSFEE882 pKa = 5.13LDD884 pKa = 3.71VSSDD888 pKa = 3.62DD889 pKa = 4.37ADD891 pKa = 3.48EE892 pKa = 4.38GTYY895 pKa = 11.13DD896 pKa = 3.64LTVSGTATAAPGFSQAIAPGTILADD921 pKa = 3.5GTATVTFTIDD931 pKa = 2.78NSANATAATSLDD943 pKa = 3.75FSNSLPAGVEE953 pKa = 4.17VAATPNAATSCTGGTLTAVAGSGTLSYY980 pKa = 10.71TGGTVGAGASCTVQADD996 pKa = 3.68VTAGTDD1002 pKa = 3.41GSYY1005 pKa = 11.35SNTSGDD1011 pKa = 3.75LTSSLGNSGSASASLTVASPEE1032 pKa = 3.83IDD1034 pKa = 3.57LQRR1037 pKa = 11.84PAATSLADD1045 pKa = 4.06GDD1047 pKa = 3.9TDD1049 pKa = 3.52AQGNVAVGVQQVLTYY1064 pKa = 9.6TVEE1067 pKa = 4.15NTGTATLTLSGTPSSAAASNVSVDD1091 pKa = 4.28SISAPGSSSLAASASTTFTVAYY1113 pKa = 8.05TPTAAGAFSFEE1124 pKa = 5.13LDD1126 pKa = 3.71VSSDD1130 pKa = 3.62DD1131 pKa = 4.37ADD1133 pKa = 3.48EE1134 pKa = 4.38GTYY1137 pKa = 11.13DD1138 pKa = 3.64LTVSGTADD1146 pKa = 3.34GSAEE1150 pKa = 4.0IGVGSATGGSLEE1162 pKa = 5.79DD1163 pKa = 4.69GDD1165 pKa = 4.71TDD1167 pKa = 4.78TIPGTPSAGSPTVIIYY1183 pKa = 7.5TFTNSGSGTLNITTPTVAGNISNEE1207 pKa = 4.13TNVVVNSLTLASNTVAGGGGTTTLEE1232 pKa = 3.54ISYY1235 pKa = 8.8TPQVSGAYY1243 pKa = 10.05RR1244 pKa = 11.84FDD1246 pKa = 3.66LQVANDD1252 pKa = 4.4DD1253 pKa = 3.9ADD1255 pKa = 3.46EE1256 pKa = 4.33SLFNITVAGSASGLPEE1272 pKa = 4.42IEE1274 pKa = 4.43VSSSASGALSDD1285 pKa = 5.17GDD1287 pKa = 3.73TDD1289 pKa = 5.08SIATVATPGVTASVTYY1305 pKa = 9.93TITNSGTDD1313 pKa = 3.74VLTLDD1318 pKa = 4.26APSVASNIRR1327 pKa = 11.84NQSNVTVDD1335 pKa = 3.68RR1336 pKa = 11.84LSLASTTVASGGGTTTLRR1354 pKa = 11.84VDD1356 pKa = 3.53YY1357 pKa = 8.79TATAGGAYY1365 pKa = 10.11GFDD1368 pKa = 3.94LSLGSDD1374 pKa = 4.38DD1375 pKa = 5.62ADD1377 pKa = 3.5EE1378 pKa = 5.14DD1379 pKa = 3.96PFDD1382 pKa = 3.77IVVSGTAQSVASALAAQSGSGQVSEE1407 pKa = 5.29VGAQFPAPLVVLVTDD1422 pKa = 3.64STGTGVAGVDD1432 pKa = 3.36VTFAAPASGASVVFASTGTHH1452 pKa = 5.82TEE1454 pKa = 4.15TVTTGSDD1461 pKa = 3.31GTASTSAMTANGTPSSYY1478 pKa = 11.34AGGSTLQSYY1487 pKa = 10.87DD1488 pKa = 3.22VTASATGLASVTFALTNDD1506 pKa = 3.81RR1507 pKa = 11.84DD1508 pKa = 3.95SEE1510 pKa = 4.18ADD1512 pKa = 3.08IQKK1515 pKa = 7.76TQEE1518 pKa = 3.77VIAAFVSNRR1527 pKa = 11.84ANAIVSNQPDD1537 pKa = 3.74LVSRR1541 pKa = 11.84LRR1543 pKa = 11.84QGALAGQQGRR1553 pKa = 11.84NGFDD1557 pKa = 3.13LRR1559 pKa = 11.84ATSGGRR1565 pKa = 11.84SANFSFSLRR1574 pKa = 11.84ALLEE1578 pKa = 3.74EE1579 pKa = 3.91ARR1581 pKa = 11.84RR1582 pKa = 11.84RR1583 pKa = 11.84NDD1585 pKa = 3.03GARR1588 pKa = 11.84WRR1590 pKa = 11.84EE1591 pKa = 3.99TLRR1594 pKa = 11.84HH1595 pKa = 6.08ADD1597 pKa = 3.27ASAGPGADD1605 pKa = 3.25SAADD1609 pKa = 3.42RR1610 pKa = 11.84AMASQAYY1617 pKa = 9.3ALRR1620 pKa = 11.84FRR1622 pKa = 11.84GTDD1625 pKa = 3.38PAQAAPVVTPVISSGWDD1642 pKa = 3.39FWAEE1646 pKa = 3.4GTYY1649 pKa = 10.55AITEE1653 pKa = 4.2SGNFEE1658 pKa = 4.15SRR1660 pKa = 11.84SGLFFAGADD1669 pKa = 3.93YY1670 pKa = 10.56RR1671 pKa = 11.84WSDD1674 pKa = 3.63QVLLGVMGQLDD1685 pKa = 3.72IADD1688 pKa = 4.48EE1689 pKa = 4.76DD1690 pKa = 3.89NVAAGTSASGTGWMVGPYY1708 pKa = 9.93VVVRR1712 pKa = 11.84LDD1714 pKa = 3.18QNLYY1718 pKa = 10.34LDD1720 pKa = 3.63VAATYY1725 pKa = 10.14GRR1727 pKa = 11.84SSNTVNALGLFEE1739 pKa = 6.72DD1740 pKa = 5.12DD1741 pKa = 4.4FDD1743 pKa = 4.21TEE1745 pKa = 4.43RR1746 pKa = 11.84FLLQGGLTGDD1756 pKa = 4.25FKK1758 pKa = 11.72LNARR1762 pKa = 11.84TTISPFARR1770 pKa = 11.84LTYY1773 pKa = 10.12YY1774 pKa = 10.31YY1775 pKa = 9.6EE1776 pKa = 4.23KK1777 pKa = 10.59QEE1779 pKa = 4.49SYY1781 pKa = 10.99TDD1783 pKa = 3.27TLGRR1787 pKa = 11.84VIPSQDD1793 pKa = 3.07FEE1795 pKa = 4.64LGRR1798 pKa = 11.84FEE1800 pKa = 4.84FGPKK1804 pKa = 9.75ISWEE1808 pKa = 4.16MYY1810 pKa = 9.38LQEE1813 pKa = 4.36GTQFSPYY1820 pKa = 10.25VSFSGIYY1827 pKa = 10.21DD1828 pKa = 3.59FNKK1831 pKa = 10.39LQGATPTDD1839 pKa = 3.28ATLASSQSDD1848 pKa = 3.24LRR1850 pKa = 11.84GRR1852 pKa = 11.84LEE1854 pKa = 4.12AGATFLVPDD1863 pKa = 4.45RR1864 pKa = 11.84GIKK1867 pKa = 9.53VAVEE1871 pKa = 3.91GFYY1874 pKa = 11.15DD1875 pKa = 5.3GIGTSDD1881 pKa = 3.76FEE1883 pKa = 4.93SYY1885 pKa = 10.6GASLSVLIPFF1895 pKa = 5.19
Molecular weight: 188.02 kDa
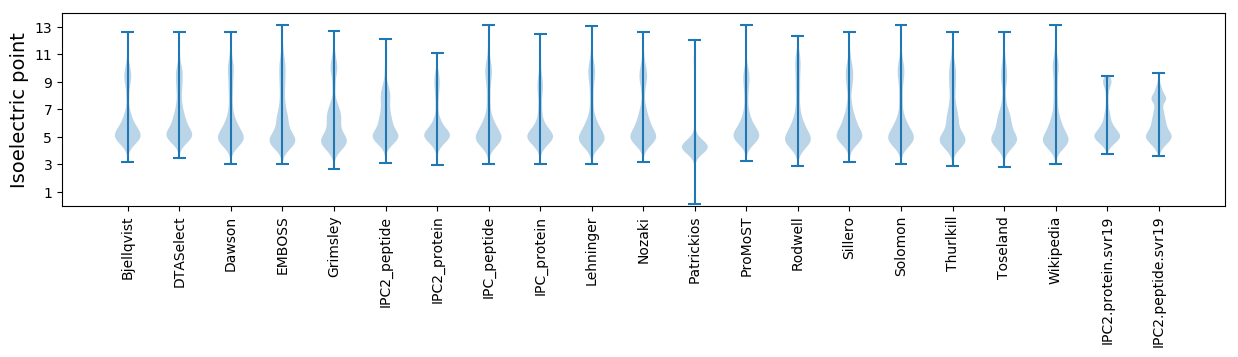
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q1GJX8|ENGB_RUEST Probable GTP-binding protein EngB OS=Ruegeria sp. (strain TM1040) OX=292414 GN=engB PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1216663 |
41 |
2150 |
316.1 |
34.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.047 ± 0.053 | 0.922 ± 0.013 |
5.95 ± 0.037 | 6.295 ± 0.039 |
3.706 ± 0.024 | 8.37 ± 0.042 |
2.092 ± 0.021 | 5.091 ± 0.025 |
3.311 ± 0.031 | 10.108 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.788 ± 0.023 | 2.657 ± 0.024 |
4.897 ± 0.03 | 3.477 ± 0.022 |
6.512 ± 0.043 | 5.536 ± 0.034 |
5.467 ± 0.034 | 7.14 ± 0.033 |
1.365 ± 0.017 | 2.27 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
