
Torque teno tupaia virus (isolate Tbc-TTV14)
Taxonomy: Viruses; Anelloviridae; Deltatorquevirus; Torque teno tupaia virus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
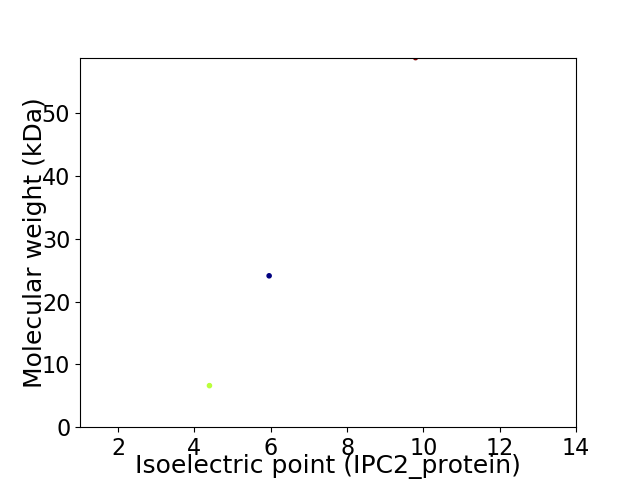
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q91PQ3|ORF3_TTVD1 Uncharacterized ORF3 protein OS=Torque teno tupaia virus (isolate Tbc-TTV14) OX=766185 GN=ORF3 PE=4 SV=1
MM1 pKa = 8.05DD2 pKa = 5.25PAKK5 pKa = 10.17IWWHH9 pKa = 5.7SCLLSHH15 pKa = 7.52KK16 pKa = 9.42SWCNCTEE23 pKa = 4.0PRR25 pKa = 11.84NHH27 pKa = 6.6LPGWPTSEE35 pKa = 4.1GTSTEE40 pKa = 4.21DD41 pKa = 3.89GDD43 pKa = 4.53IITDD47 pKa = 3.68AEE49 pKa = 4.36MLTLAEE55 pKa = 4.35DD56 pKa = 4.12TEE58 pKa = 4.79GG59 pKa = 3.74
MM1 pKa = 8.05DD2 pKa = 5.25PAKK5 pKa = 10.17IWWHH9 pKa = 5.7SCLLSHH15 pKa = 7.52KK16 pKa = 9.42SWCNCTEE23 pKa = 4.0PRR25 pKa = 11.84NHH27 pKa = 6.6LPGWPTSEE35 pKa = 4.1GTSTEE40 pKa = 4.21DD41 pKa = 3.89GDD43 pKa = 4.53IITDD47 pKa = 3.68AEE49 pKa = 4.36MLTLAEE55 pKa = 4.35DD56 pKa = 4.12TEE58 pKa = 4.79GG59 pKa = 3.74
Molecular weight: 6.62 kDa
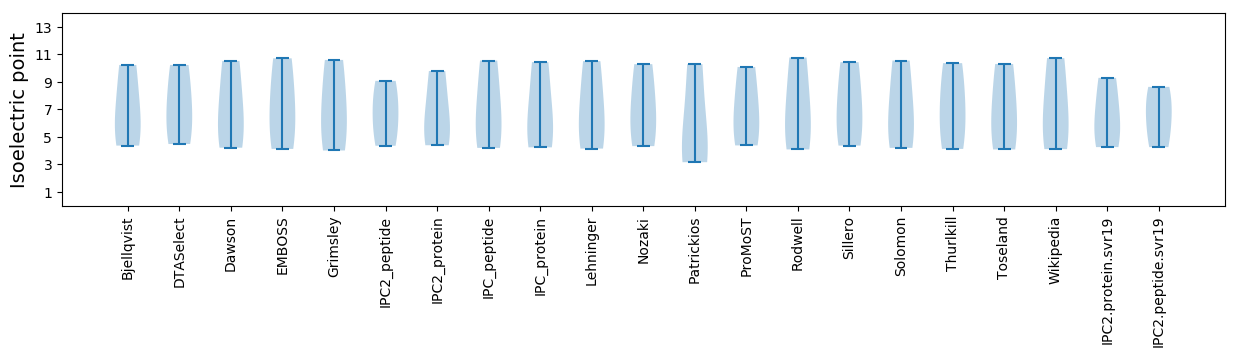
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q91PQ2|ORF2_TTVD1 Uncharacterized ORF2 protein OS=Torque teno tupaia virus (isolate Tbc-TTV14) OX=766185 GN=ORF2 PE=4 SV=1
MM1 pKa = 7.8AYY3 pKa = 10.17FRR5 pKa = 11.84RR6 pKa = 11.84NFYY9 pKa = 10.19GGRR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.81YY15 pKa = 10.39YY16 pKa = 9.29RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84NAYY22 pKa = 5.75TRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 8.99RR28 pKa = 11.84RR29 pKa = 11.84VRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84HH34 pKa = 5.42RR35 pKa = 11.84GRR37 pKa = 11.84RR38 pKa = 11.84FPYY41 pKa = 9.3FKK43 pKa = 9.85QGKK46 pKa = 8.15RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84TRR51 pKa = 11.84LLHH54 pKa = 5.08VTDD57 pKa = 4.27PRR59 pKa = 11.84YY60 pKa = 9.69KK61 pKa = 10.0AHH63 pKa = 6.11CTITGWVPIAMTRR76 pKa = 11.84IQLISQPFTVDD87 pKa = 2.83TLSPFHH93 pKa = 6.91VYY95 pKa = 10.08PGGYY99 pKa = 9.63GYY101 pKa = 11.39GVFTLEE107 pKa = 4.6AMYY110 pKa = 10.43KK111 pKa = 8.03EE112 pKa = 4.4HH113 pKa = 7.0LMKK116 pKa = 10.58RR117 pKa = 11.84NRR119 pKa = 11.84WSRR122 pKa = 11.84SNAGFDD128 pKa = 3.15LGRR131 pKa = 11.84YY132 pKa = 8.78LGTWLTLVPHH142 pKa = 7.24PYY144 pKa = 10.03FSYY147 pKa = 10.79LFYY150 pKa = 11.04YY151 pKa = 10.38DD152 pKa = 4.16PEE154 pKa = 4.62YY155 pKa = 11.47GNTFEE160 pKa = 4.46FKK162 pKa = 10.74KK163 pKa = 10.37LIHH166 pKa = 5.88PAIMITHH173 pKa = 7.66PKK175 pKa = 8.94TILVLSNKK183 pKa = 10.03RR184 pKa = 11.84GGNRR188 pKa = 11.84RR189 pKa = 11.84KK190 pKa = 9.1WPRR193 pKa = 11.84MWIPRR198 pKa = 11.84PAIFADD204 pKa = 3.34GWEE207 pKa = 4.24FQKK210 pKa = 10.85DD211 pKa = 3.66LCQHH215 pKa = 5.86GLFAFGWAWVDD226 pKa = 4.76LDD228 pKa = 4.67RR229 pKa = 11.84PWMADD234 pKa = 2.53ISKK237 pKa = 9.23PAQTIDD243 pKa = 3.06PSTYY247 pKa = 10.48DD248 pKa = 4.11KK249 pKa = 11.33KK250 pKa = 11.1NDD252 pKa = 3.96MINKK256 pKa = 8.11VPNMWWKK263 pKa = 10.13EE264 pKa = 3.8AANDD268 pKa = 3.64WLTEE272 pKa = 3.7WGRR275 pKa = 11.84PQGSGVTQQFNQLAMAGPFVLKK297 pKa = 9.68TNKK300 pKa = 9.64SEE302 pKa = 4.21YY303 pKa = 11.02VNTEE307 pKa = 3.8LLQIILFYY315 pKa = 10.58KK316 pKa = 10.16SHH318 pKa = 5.83FQWGGEE324 pKa = 3.89FLQDD328 pKa = 4.64KK329 pKa = 10.45VLADD333 pKa = 3.65PTKK336 pKa = 10.45IPPAQTAYY344 pKa = 10.07YY345 pKa = 8.95QQAGQTNLYY354 pKa = 9.06GSPYY358 pKa = 9.79KK359 pKa = 10.11PLSSGLHH366 pKa = 6.19DD367 pKa = 5.63SISEE371 pKa = 4.11LPIASPPQNPHH382 pKa = 6.79KK383 pKa = 10.01YY384 pKa = 7.14TVRR387 pKa = 11.84PSDD390 pKa = 3.54YY391 pKa = 10.72DD392 pKa = 3.21KK393 pKa = 11.7DD394 pKa = 4.3GILTEE399 pKa = 4.06EE400 pKa = 4.24SFRR403 pKa = 11.84RR404 pKa = 11.84LTDD407 pKa = 3.09SPYY410 pKa = 10.32TSDD413 pKa = 3.57GGHH416 pKa = 6.03SFSSFSTASDD426 pKa = 3.63PLEE429 pKa = 4.31GTSRR433 pKa = 11.84YY434 pKa = 9.61ARR436 pKa = 11.84PLGRR440 pKa = 11.84KK441 pKa = 4.56TTRR444 pKa = 11.84EE445 pKa = 3.31PSRR448 pKa = 11.84RR449 pKa = 11.84RR450 pKa = 11.84KK451 pKa = 9.21RR452 pKa = 11.84RR453 pKa = 11.84RR454 pKa = 11.84RR455 pKa = 11.84SPSPEE460 pKa = 4.09DD461 pKa = 3.91EE462 pKa = 4.22EE463 pKa = 4.47TAPIPPKK470 pKa = 10.75SNSEE474 pKa = 3.9PSISGPGRR482 pKa = 11.84ATRR485 pKa = 11.84KK486 pKa = 9.44QLLQRR491 pKa = 11.84LFRR494 pKa = 11.84RR495 pKa = 11.84LLTSSPQQ502 pKa = 3.05
MM1 pKa = 7.8AYY3 pKa = 10.17FRR5 pKa = 11.84RR6 pKa = 11.84NFYY9 pKa = 10.19GGRR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.81YY15 pKa = 10.39YY16 pKa = 9.29RR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84NAYY22 pKa = 5.75TRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 8.99RR28 pKa = 11.84RR29 pKa = 11.84VRR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84HH34 pKa = 5.42RR35 pKa = 11.84GRR37 pKa = 11.84RR38 pKa = 11.84FPYY41 pKa = 9.3FKK43 pKa = 9.85QGKK46 pKa = 8.15RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84TRR51 pKa = 11.84LLHH54 pKa = 5.08VTDD57 pKa = 4.27PRR59 pKa = 11.84YY60 pKa = 9.69KK61 pKa = 10.0AHH63 pKa = 6.11CTITGWVPIAMTRR76 pKa = 11.84IQLISQPFTVDD87 pKa = 2.83TLSPFHH93 pKa = 6.91VYY95 pKa = 10.08PGGYY99 pKa = 9.63GYY101 pKa = 11.39GVFTLEE107 pKa = 4.6AMYY110 pKa = 10.43KK111 pKa = 8.03EE112 pKa = 4.4HH113 pKa = 7.0LMKK116 pKa = 10.58RR117 pKa = 11.84NRR119 pKa = 11.84WSRR122 pKa = 11.84SNAGFDD128 pKa = 3.15LGRR131 pKa = 11.84YY132 pKa = 8.78LGTWLTLVPHH142 pKa = 7.24PYY144 pKa = 10.03FSYY147 pKa = 10.79LFYY150 pKa = 11.04YY151 pKa = 10.38DD152 pKa = 4.16PEE154 pKa = 4.62YY155 pKa = 11.47GNTFEE160 pKa = 4.46FKK162 pKa = 10.74KK163 pKa = 10.37LIHH166 pKa = 5.88PAIMITHH173 pKa = 7.66PKK175 pKa = 8.94TILVLSNKK183 pKa = 10.03RR184 pKa = 11.84GGNRR188 pKa = 11.84RR189 pKa = 11.84KK190 pKa = 9.1WPRR193 pKa = 11.84MWIPRR198 pKa = 11.84PAIFADD204 pKa = 3.34GWEE207 pKa = 4.24FQKK210 pKa = 10.85DD211 pKa = 3.66LCQHH215 pKa = 5.86GLFAFGWAWVDD226 pKa = 4.76LDD228 pKa = 4.67RR229 pKa = 11.84PWMADD234 pKa = 2.53ISKK237 pKa = 9.23PAQTIDD243 pKa = 3.06PSTYY247 pKa = 10.48DD248 pKa = 4.11KK249 pKa = 11.33KK250 pKa = 11.1NDD252 pKa = 3.96MINKK256 pKa = 8.11VPNMWWKK263 pKa = 10.13EE264 pKa = 3.8AANDD268 pKa = 3.64WLTEE272 pKa = 3.7WGRR275 pKa = 11.84PQGSGVTQQFNQLAMAGPFVLKK297 pKa = 9.68TNKK300 pKa = 9.64SEE302 pKa = 4.21YY303 pKa = 11.02VNTEE307 pKa = 3.8LLQIILFYY315 pKa = 10.58KK316 pKa = 10.16SHH318 pKa = 5.83FQWGGEE324 pKa = 3.89FLQDD328 pKa = 4.64KK329 pKa = 10.45VLADD333 pKa = 3.65PTKK336 pKa = 10.45IPPAQTAYY344 pKa = 10.07YY345 pKa = 8.95QQAGQTNLYY354 pKa = 9.06GSPYY358 pKa = 9.79KK359 pKa = 10.11PLSSGLHH366 pKa = 6.19DD367 pKa = 5.63SISEE371 pKa = 4.11LPIASPPQNPHH382 pKa = 6.79KK383 pKa = 10.01YY384 pKa = 7.14TVRR387 pKa = 11.84PSDD390 pKa = 3.54YY391 pKa = 10.72DD392 pKa = 3.21KK393 pKa = 11.7DD394 pKa = 4.3GILTEE399 pKa = 4.06EE400 pKa = 4.24SFRR403 pKa = 11.84RR404 pKa = 11.84LTDD407 pKa = 3.09SPYY410 pKa = 10.32TSDD413 pKa = 3.57GGHH416 pKa = 6.03SFSSFSTASDD426 pKa = 3.63PLEE429 pKa = 4.31GTSRR433 pKa = 11.84YY434 pKa = 9.61ARR436 pKa = 11.84PLGRR440 pKa = 11.84KK441 pKa = 4.56TTRR444 pKa = 11.84EE445 pKa = 3.31PSRR448 pKa = 11.84RR449 pKa = 11.84RR450 pKa = 11.84KK451 pKa = 9.21RR452 pKa = 11.84RR453 pKa = 11.84RR454 pKa = 11.84RR455 pKa = 11.84SPSPEE460 pKa = 4.09DD461 pKa = 3.91EE462 pKa = 4.22EE463 pKa = 4.47TAPIPPKK470 pKa = 10.75SNSEE474 pKa = 3.9PSISGPGRR482 pKa = 11.84ATRR485 pKa = 11.84KK486 pKa = 9.44QLLQRR491 pKa = 11.84LFRR494 pKa = 11.84RR495 pKa = 11.84LLTSSPQQ502 pKa = 3.05
Molecular weight: 58.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
775 |
59 |
502 |
258.3 |
29.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.935 ± 0.852 | 1.161 ± 0.908 |
5.677 ± 1.101 | 4.774 ± 1.211 |
3.613 ± 1.378 | 5.806 ± 1.079 |
3.484 ± 0.694 | 3.871 ± 0.454 |
5.032 ± 0.558 | 8.0 ± 0.429 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.194 ± 0.258 | 3.226 ± 0.18 |
8.387 ± 0.313 | 3.871 ± 0.708 |
9.161 ± 1.825 | 8.0 ± 0.84 |
8.258 ± 1.488 | 2.323 ± 0.698 |
2.968 ± 0.692 | 4.258 ± 1.54 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
