
Flavobacterium gilvum
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
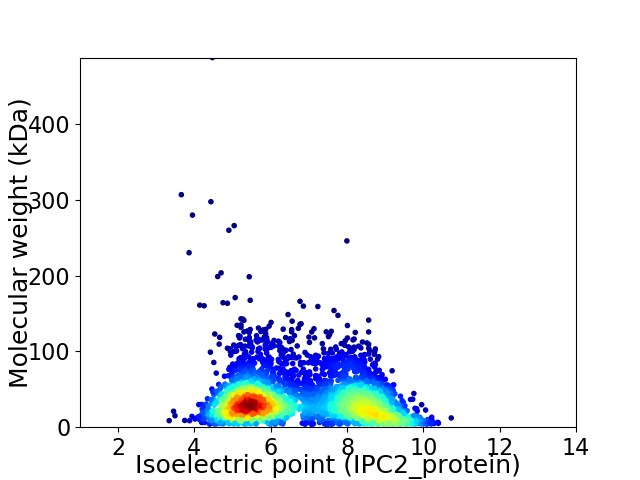
Virtual 2D-PAGE plot for 3653 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
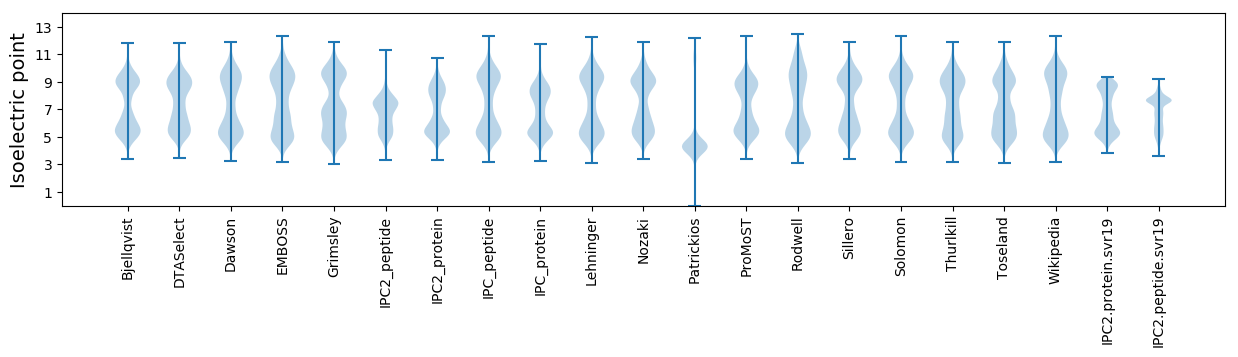
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
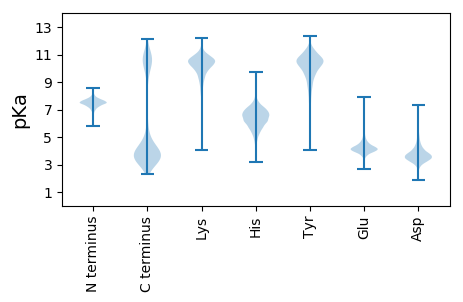
Protein with the lowest isoelectric point:
>tr|A0A085ELD3|A0A085ELD3_9FLAO Uncharacterized protein OS=Flavobacterium gilvum OX=1492737 GN=FEM08_12070 PE=4 SV=1
MM1 pKa = 7.28EE2 pKa = 5.45KK3 pKa = 10.15KK4 pKa = 10.57YY5 pKa = 10.95PNLPFNLTVFSIVSSQALFKK25 pKa = 10.75RR26 pKa = 11.84ILLLMTFYY34 pKa = 11.08LCSTTIISAQCNSDD48 pKa = 3.36VPIPFIEE55 pKa = 4.45STFGGLTATSSISGVCVCTALNEE78 pKa = 4.06PRR80 pKa = 11.84LIDD83 pKa = 4.12ANLNNFATAVVAVGAIGTEE102 pKa = 4.06NLRR105 pKa = 11.84VTDD108 pKa = 3.63TNTDD112 pKa = 3.74YY113 pKa = 11.56LAGTYY118 pKa = 10.43AGFKK122 pKa = 10.29INTGGLLNVNLLGSITITTYY142 pKa = 11.15LNGVEE147 pKa = 4.37KK148 pKa = 10.73EE149 pKa = 4.5SVTGASLLGVGLLNGSSNDD168 pKa = 3.42YY169 pKa = 10.92VVGFNTKK176 pKa = 9.15QSFDD180 pKa = 4.03AIEE183 pKa = 4.04ISVGGLVSLLNTVNVYY199 pKa = 10.63YY200 pKa = 9.82PVIRR204 pKa = 11.84NYY206 pKa = 10.51CAGPALICNTATAMNLPVYY225 pKa = 9.38PVSLEE230 pKa = 3.89PSHH233 pKa = 6.45TGVSGVSIGNIYY245 pKa = 10.72DD246 pKa = 3.64GEE248 pKa = 4.32NVISSNTNDD257 pKa = 3.31FATISLTAGVLATGNISVKK276 pKa = 10.63DD277 pKa = 3.66QLTDD281 pKa = 3.7YY282 pKa = 10.98PVGTYY287 pKa = 10.65AGFEE291 pKa = 4.09IEE293 pKa = 4.65NTSLLNVSALGNVTVTTYY311 pKa = 11.41LDD313 pKa = 3.78GVAKK317 pKa = 10.19QIISGNNLVATGTLLNSSGRR337 pKa = 11.84YY338 pKa = 8.49KK339 pKa = 10.9VGFVSTTSFDD349 pKa = 3.54EE350 pKa = 4.15VQISINQTLGVNLGSTKK367 pKa = 10.31VYY369 pKa = 10.44GAVFEE374 pKa = 4.57NFCAGPDD381 pKa = 3.82LVCNTQTGMVAPTYY395 pKa = 9.76PVYY398 pKa = 10.8INAANTGIEE407 pKa = 4.24GVACLLCSVKK417 pKa = 10.56NAEE420 pKa = 4.21NLIDD424 pKa = 3.84QTPSNYY430 pKa = 10.46AEE432 pKa = 4.19INLTAGVLASGSISVKK448 pKa = 10.59DD449 pKa = 3.58QIKK452 pKa = 10.18DD453 pKa = 3.52YY454 pKa = 10.6PIGSFVGFTVEE465 pKa = 4.59NPALLSVNALDD476 pKa = 5.6AITITTYY483 pKa = 11.3LDD485 pKa = 3.57GVPQEE490 pKa = 4.12IKK492 pKa = 10.8AGNTALATVGTNLLVGTSPQTIGFIATKK520 pKa = 10.4SFDD523 pKa = 3.47EE524 pKa = 4.36ARR526 pKa = 11.84ITIANTASVNLGIVRR541 pKa = 11.84VYY543 pKa = 10.31QAVFEE548 pKa = 4.36KK549 pKa = 10.59LCPVTLEE556 pKa = 4.14CNKK559 pKa = 9.93TYY561 pKa = 11.18SLTNPTFPVIIDD573 pKa = 3.65SNKK576 pKa = 9.34TGVQGVACVSCAVNDD591 pKa = 4.07TNNVLTASTTDD602 pKa = 3.73FARR605 pKa = 11.84IMVTAGVIAPASIAVLDD622 pKa = 3.89QLSTYY627 pKa = 9.89PAGTIAGFTIRR638 pKa = 11.84DD639 pKa = 3.89RR640 pKa = 11.84NSLLQLDD647 pKa = 5.08LFNSLTISTYY657 pKa = 10.89LDD659 pKa = 3.22GVLQEE664 pKa = 4.82SRR666 pKa = 11.84SASNIINLNLLTSFGTGAGMYY687 pKa = 9.99NVGFRR692 pKa = 11.84ATKK695 pKa = 10.18SFDD698 pKa = 3.65EE699 pKa = 3.98IRR701 pKa = 11.84ITVGSLASVINDD713 pKa = 2.94INVYY717 pKa = 9.87GAFVNTSGTSGGTLFCAVIDD737 pKa = 5.21AIDD740 pKa = 4.45DD741 pKa = 3.84NFPPIAGASGDD752 pKa = 4.11SNIGNVLTNDD762 pKa = 3.78TLSGNPATISDD773 pKa = 3.92VNLAVLTPAVPLTVGASVPSIDD795 pKa = 3.2IATGVVSVPAGTPAGNYY812 pKa = 9.75SITYY816 pKa = 7.04QICYY820 pKa = 9.64KK821 pKa = 10.56LNSLTCDD828 pKa = 3.03TATVNIVVTAATINAEE844 pKa = 4.2DD845 pKa = 4.2DD846 pKa = 4.01TNTTTIYY853 pKa = 10.98GNVGGNPGINVYY865 pKa = 11.18ANDD868 pKa = 3.86TTVGGVLINPSDD880 pKa = 3.87VTLSSTPNGPLTVNADD896 pKa = 3.36GSVTVAPNTPAGTYY910 pKa = 7.77TVTYY914 pKa = 7.35TICEE918 pKa = 4.07NLNGSNCDD926 pKa = 3.46SATVTVLVSAVPIDD940 pKa = 4.25AVNDD944 pKa = 3.77NNNTAPIIYY953 pKa = 10.21GNVGGNAGINVYY965 pKa = 11.11ANDD968 pKa = 3.94TLNGVLVNPSDD979 pKa = 4.1VIFGSSPNGPLTVNADD995 pKa = 3.36GSVTVAPNTPAGTYY1009 pKa = 8.32TVDD1012 pKa = 3.25YY1013 pKa = 7.94TICEE1017 pKa = 4.35KK1018 pKa = 11.02LNSANNCDD1026 pKa = 3.63TATVTVVVSAAVVDD1040 pKa = 4.21AVVDD1044 pKa = 4.06TNTLPVYY1051 pKa = 10.89GNVGGNTDD1059 pKa = 3.15INVYY1063 pKa = 11.11ANDD1066 pKa = 3.88TLNGVLINPSDD1077 pKa = 3.74FTLISTPTSILTINPDD1093 pKa = 2.85GSVTVAPKK1101 pKa = 9.98TPAGTYY1107 pKa = 9.92SVDD1110 pKa = 3.3YY1111 pKa = 8.47TICEE1115 pKa = 4.35KK1116 pKa = 11.06LNSTNNCDD1124 pKa = 3.37TATVTVVVSKK1134 pKa = 11.02AVVDD1138 pKa = 4.01AVVDD1142 pKa = 4.06TNTLPVYY1149 pKa = 10.89GNVGGNTDD1157 pKa = 3.51INVLANDD1164 pKa = 4.06TQNGVPVNRR1173 pKa = 11.84LDD1175 pKa = 3.55VTLTSTPTSILTINPDD1191 pKa = 2.85GSVTVAPNTPAGSYY1205 pKa = 9.59TIDD1208 pKa = 3.28YY1209 pKa = 7.52TICEE1213 pKa = 4.39KK1214 pKa = 11.25LNLTTNCDD1222 pKa = 3.37TATVTVVVSAAVVDD1236 pKa = 4.21AVVDD1240 pKa = 4.06TNTLPVYY1247 pKa = 10.89GNVGGNTDD1255 pKa = 3.15INVYY1259 pKa = 11.11ANDD1262 pKa = 3.88TLNGVLINPSDD1273 pKa = 3.86FTLTSTPTSQLKK1285 pKa = 10.62VNTDD1289 pKa = 3.03GSVTVAPNTPVGTYY1303 pKa = 10.2SVDD1306 pKa = 3.32YY1307 pKa = 8.46TICEE1311 pKa = 4.2KK1312 pKa = 11.25LNLTTNCDD1320 pKa = 3.37TATVTVVVSAAVVDD1334 pKa = 4.21AVVDD1338 pKa = 4.06TNTLPVYY1345 pKa = 10.89GNVGGNTGINVLANDD1360 pKa = 4.13TQNGVPVNPLDD1371 pKa = 3.52VTLISTPTSILTINPDD1387 pKa = 2.86GSVTVASNTPVGTYY1401 pKa = 10.29SVDD1404 pKa = 3.32YY1405 pKa = 8.46TICEE1409 pKa = 4.3KK1410 pKa = 11.08LNSTTNCDD1418 pKa = 3.28TATVTVVVSAAVVDD1432 pKa = 4.21AVVDD1436 pKa = 4.06TNTLPVYY1443 pKa = 10.89GNVGGNTGINVLANDD1458 pKa = 4.13TQNGVPVNPLDD1469 pKa = 3.68VTLTSTPNGPLTINPDD1485 pKa = 2.98GSVTVAPNTPAGTYY1499 pKa = 10.05SVDD1502 pKa = 3.3YY1503 pKa = 8.47TICEE1507 pKa = 4.3KK1508 pKa = 11.08LNSTTNCDD1516 pKa = 3.27TATVTVAVSFPSIAVVKK1533 pKa = 8.59TSVVGGTGKK1542 pKa = 10.75VGDD1545 pKa = 3.97LVTYY1549 pKa = 8.32TFTVTNTGDD1558 pKa = 3.24TTLNGIVINDD1568 pKa = 3.71ALTNSVNLSVTPSTLVPNGIGTATSTYY1595 pKa = 9.96IIKK1598 pKa = 10.61QSDD1601 pKa = 2.96IDD1603 pKa = 3.62AGKK1606 pKa = 8.19ITNSATVTGTIPTGGTVTDD1625 pKa = 3.92ISGTTITDD1633 pKa = 3.22NTPTVTTLTTSPSIAVVKK1651 pKa = 9.36TSVVGGTGKK1660 pKa = 10.74VGDD1663 pKa = 3.86VVTYY1667 pKa = 9.34TFTVTNTGNTTLSNIVINDD1686 pKa = 3.31VLTNTVNKK1694 pKa = 9.79VVTVGNLAPNATATTTVTYY1713 pKa = 10.13IIKK1716 pKa = 10.54QSDD1719 pKa = 2.96IDD1721 pKa = 3.62AGKK1724 pKa = 8.19ITNSATVTGTTPAGGTVTDD1743 pKa = 4.01ISGTAITNDD1752 pKa = 3.45TPTVTTLTTSPSIAVVKK1769 pKa = 9.35TSVVGTTIKK1778 pKa = 10.95VGDD1781 pKa = 3.66KK1782 pKa = 9.05VTYY1785 pKa = 9.55TFTVTNTGNTTLNNIVINDD1804 pKa = 3.82ALTNTVNLAVTPSTLAPNSIGTATSTYY1831 pKa = 9.96IIKK1834 pKa = 10.61QSDD1837 pKa = 3.03IDD1839 pKa = 3.95AGQITNSATVIGTIPTGGTVTDD1861 pKa = 3.8ISGTAVNNDD1870 pKa = 3.04NPTITTLNTNSAISIMKK1887 pKa = 9.99EE1888 pKa = 3.37GTYY1891 pKa = 10.24EE1892 pKa = 4.16DD1893 pKa = 4.22KK1894 pKa = 11.51NKK1896 pKa = 11.07DD1897 pKa = 3.22GIANVGDD1904 pKa = 3.74VINYY1908 pKa = 9.27SFTVTNTGNVSLHH1921 pKa = 6.67NITIQDD1927 pKa = 3.75NNVTTVKK1934 pKa = 9.64GTLSSLSVGASDD1946 pKa = 4.26SVAFSAVYY1954 pKa = 10.48SITDD1958 pKa = 3.28ADD1960 pKa = 3.61IVSGYY1965 pKa = 9.87VYY1967 pKa = 10.84NSAIISALSSANAKK1981 pKa = 7.9VTAVSTDD1988 pKa = 3.46PTPCIQCPVLSNCLTCTITAIPQAPSIAIIKK2019 pKa = 10.14KK2020 pKa = 10.02GVFRR2024 pKa = 11.84DD2025 pKa = 3.9EE2026 pKa = 4.83NGDD2029 pKa = 3.43GYY2031 pKa = 11.67AEE2033 pKa = 4.11IGEE2036 pKa = 4.45TIDD2039 pKa = 3.78YY2040 pKa = 10.73SFTIMNTGNLPLTNVTLTDD2059 pKa = 3.67ALVGIVISGGPINLAVGATDD2079 pKa = 3.24TTTFKK2084 pKa = 10.43ATYY2087 pKa = 9.6HH2088 pKa = 5.09VTQQDD2093 pKa = 3.93VVKK2096 pKa = 11.09GSVTNQAIIKK2106 pKa = 9.65GFTPTGAQVTDD2117 pKa = 4.43LSDD2120 pKa = 4.74DD2121 pKa = 4.18DD2122 pKa = 6.21SPLQDD2127 pKa = 4.46DD2128 pKa = 4.19PTVLEE2133 pKa = 4.33IDD2135 pKa = 3.69GCTLEE2140 pKa = 4.29VFNAVSPDD2148 pKa = 3.3GDD2150 pKa = 3.67GLNDD2154 pKa = 3.37IFRR2157 pKa = 11.84IRR2159 pKa = 11.84GIEE2162 pKa = 4.19CYY2164 pKa = 9.52PKK2166 pKa = 10.89NSVQVFNRR2174 pKa = 11.84WGVKK2178 pKa = 8.93VYY2180 pKa = 9.92EE2181 pKa = 4.19AEE2183 pKa = 4.55GYY2185 pKa = 11.1NNDD2188 pKa = 2.95TVAFKK2193 pKa = 10.54GYY2195 pKa = 9.93SEE2197 pKa = 4.7GRR2199 pKa = 11.84VTVNKK2204 pKa = 10.55SEE2206 pKa = 3.96GLPTGTYY2213 pKa = 9.69FYY2215 pKa = 10.57VIKK2218 pKa = 11.22YY2219 pKa = 9.82EE2220 pKa = 4.21DD2221 pKa = 3.83FSGAGIDD2228 pKa = 3.28KK2229 pKa = 10.61SGYY2232 pKa = 10.68LNISRR2237 pKa = 11.84NN2238 pKa = 3.55
MM1 pKa = 7.28EE2 pKa = 5.45KK3 pKa = 10.15KK4 pKa = 10.57YY5 pKa = 10.95PNLPFNLTVFSIVSSQALFKK25 pKa = 10.75RR26 pKa = 11.84ILLLMTFYY34 pKa = 11.08LCSTTIISAQCNSDD48 pKa = 3.36VPIPFIEE55 pKa = 4.45STFGGLTATSSISGVCVCTALNEE78 pKa = 4.06PRR80 pKa = 11.84LIDD83 pKa = 4.12ANLNNFATAVVAVGAIGTEE102 pKa = 4.06NLRR105 pKa = 11.84VTDD108 pKa = 3.63TNTDD112 pKa = 3.74YY113 pKa = 11.56LAGTYY118 pKa = 10.43AGFKK122 pKa = 10.29INTGGLLNVNLLGSITITTYY142 pKa = 11.15LNGVEE147 pKa = 4.37KK148 pKa = 10.73EE149 pKa = 4.5SVTGASLLGVGLLNGSSNDD168 pKa = 3.42YY169 pKa = 10.92VVGFNTKK176 pKa = 9.15QSFDD180 pKa = 4.03AIEE183 pKa = 4.04ISVGGLVSLLNTVNVYY199 pKa = 10.63YY200 pKa = 9.82PVIRR204 pKa = 11.84NYY206 pKa = 10.51CAGPALICNTATAMNLPVYY225 pKa = 9.38PVSLEE230 pKa = 3.89PSHH233 pKa = 6.45TGVSGVSIGNIYY245 pKa = 10.72DD246 pKa = 3.64GEE248 pKa = 4.32NVISSNTNDD257 pKa = 3.31FATISLTAGVLATGNISVKK276 pKa = 10.63DD277 pKa = 3.66QLTDD281 pKa = 3.7YY282 pKa = 10.98PVGTYY287 pKa = 10.65AGFEE291 pKa = 4.09IEE293 pKa = 4.65NTSLLNVSALGNVTVTTYY311 pKa = 11.41LDD313 pKa = 3.78GVAKK317 pKa = 10.19QIISGNNLVATGTLLNSSGRR337 pKa = 11.84YY338 pKa = 8.49KK339 pKa = 10.9VGFVSTTSFDD349 pKa = 3.54EE350 pKa = 4.15VQISINQTLGVNLGSTKK367 pKa = 10.31VYY369 pKa = 10.44GAVFEE374 pKa = 4.57NFCAGPDD381 pKa = 3.82LVCNTQTGMVAPTYY395 pKa = 9.76PVYY398 pKa = 10.8INAANTGIEE407 pKa = 4.24GVACLLCSVKK417 pKa = 10.56NAEE420 pKa = 4.21NLIDD424 pKa = 3.84QTPSNYY430 pKa = 10.46AEE432 pKa = 4.19INLTAGVLASGSISVKK448 pKa = 10.59DD449 pKa = 3.58QIKK452 pKa = 10.18DD453 pKa = 3.52YY454 pKa = 10.6PIGSFVGFTVEE465 pKa = 4.59NPALLSVNALDD476 pKa = 5.6AITITTYY483 pKa = 11.3LDD485 pKa = 3.57GVPQEE490 pKa = 4.12IKK492 pKa = 10.8AGNTALATVGTNLLVGTSPQTIGFIATKK520 pKa = 10.4SFDD523 pKa = 3.47EE524 pKa = 4.36ARR526 pKa = 11.84ITIANTASVNLGIVRR541 pKa = 11.84VYY543 pKa = 10.31QAVFEE548 pKa = 4.36KK549 pKa = 10.59LCPVTLEE556 pKa = 4.14CNKK559 pKa = 9.93TYY561 pKa = 11.18SLTNPTFPVIIDD573 pKa = 3.65SNKK576 pKa = 9.34TGVQGVACVSCAVNDD591 pKa = 4.07TNNVLTASTTDD602 pKa = 3.73FARR605 pKa = 11.84IMVTAGVIAPASIAVLDD622 pKa = 3.89QLSTYY627 pKa = 9.89PAGTIAGFTIRR638 pKa = 11.84DD639 pKa = 3.89RR640 pKa = 11.84NSLLQLDD647 pKa = 5.08LFNSLTISTYY657 pKa = 10.89LDD659 pKa = 3.22GVLQEE664 pKa = 4.82SRR666 pKa = 11.84SASNIINLNLLTSFGTGAGMYY687 pKa = 9.99NVGFRR692 pKa = 11.84ATKK695 pKa = 10.18SFDD698 pKa = 3.65EE699 pKa = 3.98IRR701 pKa = 11.84ITVGSLASVINDD713 pKa = 2.94INVYY717 pKa = 9.87GAFVNTSGTSGGTLFCAVIDD737 pKa = 5.21AIDD740 pKa = 4.45DD741 pKa = 3.84NFPPIAGASGDD752 pKa = 4.11SNIGNVLTNDD762 pKa = 3.78TLSGNPATISDD773 pKa = 3.92VNLAVLTPAVPLTVGASVPSIDD795 pKa = 3.2IATGVVSVPAGTPAGNYY812 pKa = 9.75SITYY816 pKa = 7.04QICYY820 pKa = 9.64KK821 pKa = 10.56LNSLTCDD828 pKa = 3.03TATVNIVVTAATINAEE844 pKa = 4.2DD845 pKa = 4.2DD846 pKa = 4.01TNTTTIYY853 pKa = 10.98GNVGGNPGINVYY865 pKa = 11.18ANDD868 pKa = 3.86TTVGGVLINPSDD880 pKa = 3.87VTLSSTPNGPLTVNADD896 pKa = 3.36GSVTVAPNTPAGTYY910 pKa = 7.77TVTYY914 pKa = 7.35TICEE918 pKa = 4.07NLNGSNCDD926 pKa = 3.46SATVTVLVSAVPIDD940 pKa = 4.25AVNDD944 pKa = 3.77NNNTAPIIYY953 pKa = 10.21GNVGGNAGINVYY965 pKa = 11.11ANDD968 pKa = 3.94TLNGVLVNPSDD979 pKa = 4.1VIFGSSPNGPLTVNADD995 pKa = 3.36GSVTVAPNTPAGTYY1009 pKa = 8.32TVDD1012 pKa = 3.25YY1013 pKa = 7.94TICEE1017 pKa = 4.35KK1018 pKa = 11.02LNSANNCDD1026 pKa = 3.63TATVTVVVSAAVVDD1040 pKa = 4.21AVVDD1044 pKa = 4.06TNTLPVYY1051 pKa = 10.89GNVGGNTDD1059 pKa = 3.15INVYY1063 pKa = 11.11ANDD1066 pKa = 3.88TLNGVLINPSDD1077 pKa = 3.74FTLISTPTSILTINPDD1093 pKa = 2.85GSVTVAPKK1101 pKa = 9.98TPAGTYY1107 pKa = 9.92SVDD1110 pKa = 3.3YY1111 pKa = 8.47TICEE1115 pKa = 4.35KK1116 pKa = 11.06LNSTNNCDD1124 pKa = 3.37TATVTVVVSKK1134 pKa = 11.02AVVDD1138 pKa = 4.01AVVDD1142 pKa = 4.06TNTLPVYY1149 pKa = 10.89GNVGGNTDD1157 pKa = 3.51INVLANDD1164 pKa = 4.06TQNGVPVNRR1173 pKa = 11.84LDD1175 pKa = 3.55VTLTSTPTSILTINPDD1191 pKa = 2.85GSVTVAPNTPAGSYY1205 pKa = 9.59TIDD1208 pKa = 3.28YY1209 pKa = 7.52TICEE1213 pKa = 4.39KK1214 pKa = 11.25LNLTTNCDD1222 pKa = 3.37TATVTVVVSAAVVDD1236 pKa = 4.21AVVDD1240 pKa = 4.06TNTLPVYY1247 pKa = 10.89GNVGGNTDD1255 pKa = 3.15INVYY1259 pKa = 11.11ANDD1262 pKa = 3.88TLNGVLINPSDD1273 pKa = 3.86FTLTSTPTSQLKK1285 pKa = 10.62VNTDD1289 pKa = 3.03GSVTVAPNTPVGTYY1303 pKa = 10.2SVDD1306 pKa = 3.32YY1307 pKa = 8.46TICEE1311 pKa = 4.2KK1312 pKa = 11.25LNLTTNCDD1320 pKa = 3.37TATVTVVVSAAVVDD1334 pKa = 4.21AVVDD1338 pKa = 4.06TNTLPVYY1345 pKa = 10.89GNVGGNTGINVLANDD1360 pKa = 4.13TQNGVPVNPLDD1371 pKa = 3.52VTLISTPTSILTINPDD1387 pKa = 2.86GSVTVASNTPVGTYY1401 pKa = 10.29SVDD1404 pKa = 3.32YY1405 pKa = 8.46TICEE1409 pKa = 4.3KK1410 pKa = 11.08LNSTTNCDD1418 pKa = 3.28TATVTVVVSAAVVDD1432 pKa = 4.21AVVDD1436 pKa = 4.06TNTLPVYY1443 pKa = 10.89GNVGGNTGINVLANDD1458 pKa = 4.13TQNGVPVNPLDD1469 pKa = 3.68VTLTSTPNGPLTINPDD1485 pKa = 2.98GSVTVAPNTPAGTYY1499 pKa = 10.05SVDD1502 pKa = 3.3YY1503 pKa = 8.47TICEE1507 pKa = 4.3KK1508 pKa = 11.08LNSTTNCDD1516 pKa = 3.27TATVTVAVSFPSIAVVKK1533 pKa = 8.59TSVVGGTGKK1542 pKa = 10.75VGDD1545 pKa = 3.97LVTYY1549 pKa = 8.32TFTVTNTGDD1558 pKa = 3.24TTLNGIVINDD1568 pKa = 3.71ALTNSVNLSVTPSTLVPNGIGTATSTYY1595 pKa = 9.96IIKK1598 pKa = 10.61QSDD1601 pKa = 2.96IDD1603 pKa = 3.62AGKK1606 pKa = 8.19ITNSATVTGTIPTGGTVTDD1625 pKa = 3.92ISGTTITDD1633 pKa = 3.22NTPTVTTLTTSPSIAVVKK1651 pKa = 9.36TSVVGGTGKK1660 pKa = 10.74VGDD1663 pKa = 3.86VVTYY1667 pKa = 9.34TFTVTNTGNTTLSNIVINDD1686 pKa = 3.31VLTNTVNKK1694 pKa = 9.79VVTVGNLAPNATATTTVTYY1713 pKa = 10.13IIKK1716 pKa = 10.54QSDD1719 pKa = 2.96IDD1721 pKa = 3.62AGKK1724 pKa = 8.19ITNSATVTGTTPAGGTVTDD1743 pKa = 4.01ISGTAITNDD1752 pKa = 3.45TPTVTTLTTSPSIAVVKK1769 pKa = 9.35TSVVGTTIKK1778 pKa = 10.95VGDD1781 pKa = 3.66KK1782 pKa = 9.05VTYY1785 pKa = 9.55TFTVTNTGNTTLNNIVINDD1804 pKa = 3.82ALTNTVNLAVTPSTLAPNSIGTATSTYY1831 pKa = 9.96IIKK1834 pKa = 10.61QSDD1837 pKa = 3.03IDD1839 pKa = 3.95AGQITNSATVIGTIPTGGTVTDD1861 pKa = 3.8ISGTAVNNDD1870 pKa = 3.04NPTITTLNTNSAISIMKK1887 pKa = 9.99EE1888 pKa = 3.37GTYY1891 pKa = 10.24EE1892 pKa = 4.16DD1893 pKa = 4.22KK1894 pKa = 11.51NKK1896 pKa = 11.07DD1897 pKa = 3.22GIANVGDD1904 pKa = 3.74VINYY1908 pKa = 9.27SFTVTNTGNVSLHH1921 pKa = 6.67NITIQDD1927 pKa = 3.75NNVTTVKK1934 pKa = 9.64GTLSSLSVGASDD1946 pKa = 4.26SVAFSAVYY1954 pKa = 10.48SITDD1958 pKa = 3.28ADD1960 pKa = 3.61IVSGYY1965 pKa = 9.87VYY1967 pKa = 10.84NSAIISALSSANAKK1981 pKa = 7.9VTAVSTDD1988 pKa = 3.46PTPCIQCPVLSNCLTCTITAIPQAPSIAIIKK2019 pKa = 10.14KK2020 pKa = 10.02GVFRR2024 pKa = 11.84DD2025 pKa = 3.9EE2026 pKa = 4.83NGDD2029 pKa = 3.43GYY2031 pKa = 11.67AEE2033 pKa = 4.11IGEE2036 pKa = 4.45TIDD2039 pKa = 3.78YY2040 pKa = 10.73SFTIMNTGNLPLTNVTLTDD2059 pKa = 3.67ALVGIVISGGPINLAVGATDD2079 pKa = 3.24TTTFKK2084 pKa = 10.43ATYY2087 pKa = 9.6HH2088 pKa = 5.09VTQQDD2093 pKa = 3.93VVKK2096 pKa = 11.09GSVTNQAIIKK2106 pKa = 9.65GFTPTGAQVTDD2117 pKa = 4.43LSDD2120 pKa = 4.74DD2121 pKa = 4.18DD2122 pKa = 6.21SPLQDD2127 pKa = 4.46DD2128 pKa = 4.19PTVLEE2133 pKa = 4.33IDD2135 pKa = 3.69GCTLEE2140 pKa = 4.29VFNAVSPDD2148 pKa = 3.3GDD2150 pKa = 3.67GLNDD2154 pKa = 3.37IFRR2157 pKa = 11.84IRR2159 pKa = 11.84GIEE2162 pKa = 4.19CYY2164 pKa = 9.52PKK2166 pKa = 10.89NSVQVFNRR2174 pKa = 11.84WGVKK2178 pKa = 8.93VYY2180 pKa = 9.92EE2181 pKa = 4.19AEE2183 pKa = 4.55GYY2185 pKa = 11.1NNDD2188 pKa = 2.95TVAFKK2193 pKa = 10.54GYY2195 pKa = 9.93SEE2197 pKa = 4.7GRR2199 pKa = 11.84VTVNKK2204 pKa = 10.55SEE2206 pKa = 3.96GLPTGTYY2213 pKa = 9.69FYY2215 pKa = 10.57VIKK2218 pKa = 11.22YY2219 pKa = 9.82EE2220 pKa = 4.21DD2221 pKa = 3.83FSGAGIDD2228 pKa = 3.28KK2229 pKa = 10.61SGYY2232 pKa = 10.68LNISRR2237 pKa = 11.84NN2238 pKa = 3.55
Molecular weight: 230.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A085EJ68|A0A085EJ68_9FLAO Peptidase M3 OS=Flavobacterium gilvum OX=1492737 GN=FEM08_18670 PE=3 SV=1
MM1 pKa = 7.58PFWRR5 pKa = 11.84SLIIDD10 pKa = 3.76IGLYY14 pKa = 8.45ICKK17 pKa = 9.7SRR19 pKa = 11.84SVIGLGFLTVNKK31 pKa = 8.61KK32 pKa = 9.23TIVSIPDD39 pKa = 3.06SRR41 pKa = 11.84YY42 pKa = 8.83CRR44 pKa = 11.84PFFVFGNRR52 pKa = 11.84IHH54 pKa = 6.93GKK56 pKa = 9.58ASTKK60 pKa = 7.62EE61 pKa = 4.11THH63 pKa = 5.77FHH65 pKa = 6.88NSTKK69 pKa = 10.38LCRR72 pKa = 11.84ISIRR76 pKa = 11.84YY77 pKa = 7.76IQDD80 pKa = 2.82RR81 pKa = 11.84RR82 pKa = 11.84HH83 pKa = 6.26FISIFSLKK91 pKa = 9.15TSRR94 pKa = 11.84RR95 pKa = 11.84KK96 pKa = 10.02INILNQIGIGKK107 pKa = 6.26TQSFLLSRR115 pKa = 11.84SDD117 pKa = 3.52EE118 pKa = 3.68KK119 pKa = 10.99RR120 pKa = 11.84AIYY123 pKa = 9.44FYY125 pKa = 10.69FVNINQILIIISTSNSILCTQLIIGIDD152 pKa = 3.55SGQSFQNVFDD162 pKa = 3.64TGIGSRR168 pKa = 11.84HH169 pKa = 5.68KK170 pKa = 10.75PNVFGFKK177 pKa = 10.61SNNPLNCFFIGRR189 pKa = 11.84INDD192 pKa = 3.72NFSHH196 pKa = 7.05LLLIITHH203 pKa = 5.0VHH205 pKa = 5.91IKK207 pKa = 10.04NKK209 pKa = 9.92NFIWRR214 pKa = 11.84QQ215 pKa = 3.11
MM1 pKa = 7.58PFWRR5 pKa = 11.84SLIIDD10 pKa = 3.76IGLYY14 pKa = 8.45ICKK17 pKa = 9.7SRR19 pKa = 11.84SVIGLGFLTVNKK31 pKa = 8.61KK32 pKa = 9.23TIVSIPDD39 pKa = 3.06SRR41 pKa = 11.84YY42 pKa = 8.83CRR44 pKa = 11.84PFFVFGNRR52 pKa = 11.84IHH54 pKa = 6.93GKK56 pKa = 9.58ASTKK60 pKa = 7.62EE61 pKa = 4.11THH63 pKa = 5.77FHH65 pKa = 6.88NSTKK69 pKa = 10.38LCRR72 pKa = 11.84ISIRR76 pKa = 11.84YY77 pKa = 7.76IQDD80 pKa = 2.82RR81 pKa = 11.84RR82 pKa = 11.84HH83 pKa = 6.26FISIFSLKK91 pKa = 9.15TSRR94 pKa = 11.84RR95 pKa = 11.84KK96 pKa = 10.02INILNQIGIGKK107 pKa = 6.26TQSFLLSRR115 pKa = 11.84SDD117 pKa = 3.52EE118 pKa = 3.68KK119 pKa = 10.99RR120 pKa = 11.84AIYY123 pKa = 9.44FYY125 pKa = 10.69FVNINQILIIISTSNSILCTQLIIGIDD152 pKa = 3.55SGQSFQNVFDD162 pKa = 3.64TGIGSRR168 pKa = 11.84HH169 pKa = 5.68KK170 pKa = 10.75PNVFGFKK177 pKa = 10.61SNNPLNCFFIGRR189 pKa = 11.84INDD192 pKa = 3.72NFSHH196 pKa = 7.05LLLIITHH203 pKa = 5.0VHH205 pKa = 5.91IKK207 pKa = 10.04NKK209 pKa = 9.92NFIWRR214 pKa = 11.84QQ215 pKa = 3.11
Molecular weight: 24.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1237433 |
37 |
4660 |
338.7 |
38.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.42 ± 0.041 | 0.827 ± 0.015 |
5.345 ± 0.029 | 6.26 ± 0.051 |
5.267 ± 0.038 | 6.518 ± 0.045 |
1.662 ± 0.018 | 7.91 ± 0.043 |
8.237 ± 0.052 | 8.883 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.246 ± 0.022 | 6.442 ± 0.043 |
3.582 ± 0.027 | 3.364 ± 0.02 |
3.118 ± 0.028 | 6.529 ± 0.032 |
5.9 ± 0.064 | 6.253 ± 0.036 |
1.16 ± 0.017 | 4.077 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
