
Rat cytomegalovirus (isolate England) (RCMV-E) (Murid herpesvirus 8)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Betaherpesvirinae; Muromegalovirus
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
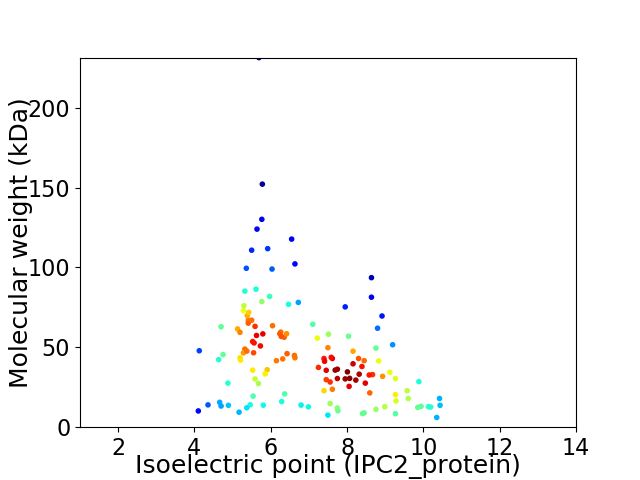
Virtual 2D-PAGE plot for 137 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|K7YA71|K7YA71_RCMVE EORF6 OS=Rat cytomegalovirus (isolate England) OX=1261657 GN=eORF6 PE=4 SV=1
MM1 pKa = 7.16TRR3 pKa = 11.84FIILSCVLCIAISSVATTMAPNVSGNDD30 pKa = 3.3MYY32 pKa = 11.29FEE34 pKa = 4.5NSGSGSSEE42 pKa = 3.74YY43 pKa = 10.74DD44 pKa = 4.04DD45 pKa = 5.13EE46 pKa = 5.39DD47 pKa = 4.35LSSGEE52 pKa = 4.14SDD54 pKa = 3.4MNSTILTEE62 pKa = 4.18NMEE65 pKa = 4.5STSNASDD72 pKa = 3.34GTDD75 pKa = 3.01EE76 pKa = 5.08YY77 pKa = 10.09NTQVTTFPNDD87 pKa = 2.76VTTDD91 pKa = 3.95FTLTPNNNYY100 pKa = 8.14STLSDD105 pKa = 3.34NRR107 pKa = 11.84SPEE110 pKa = 4.28TKK112 pKa = 10.58NPTDD116 pKa = 3.68TQTSTTPTEE125 pKa = 3.84TSISIKK131 pKa = 10.14TPTQTSAQMIQLTNTPTPTEE151 pKa = 4.1LPNTEE156 pKa = 4.38SPSLTTEE163 pKa = 3.97AVTSMKK169 pKa = 10.27TYY171 pKa = 10.29ILTSRR176 pKa = 11.84EE177 pKa = 4.09SSSKK181 pKa = 10.17SSEE184 pKa = 4.23DD185 pKa = 3.06IKK187 pKa = 11.3SEE189 pKa = 4.28NITSTSNTVYY199 pKa = 9.21KK200 pKa = 10.71TSDD203 pKa = 3.17TDD205 pKa = 3.43VFTYY209 pKa = 8.76QTTNASPTTISTEE222 pKa = 4.04TSVPTTPDD230 pKa = 3.16TKK232 pKa = 11.14SLDD235 pKa = 3.9SNKK238 pKa = 8.56VTDD241 pKa = 4.39SVTGTNSTMVTNNSSATIHH260 pKa = 6.53ASTPISHH267 pKa = 7.33TSTVVYY273 pKa = 8.7ITPHH277 pKa = 6.73NITLPDD283 pKa = 3.5TSTVSIAPHH292 pKa = 6.78NITSFDD298 pKa = 3.46TSTVSTIPHH307 pKa = 5.49VTPSDD312 pKa = 3.73YY313 pKa = 11.51NHH315 pKa = 7.12NITNHH320 pKa = 5.4NVTLYY325 pKa = 10.82DD326 pKa = 3.61YY327 pKa = 10.49DD328 pKa = 4.07YY329 pKa = 11.68KK330 pKa = 11.29NLQTTSSVSVSDD342 pKa = 4.86LYY344 pKa = 10.84ISTTIQTTPLFTPVPCISNYY364 pKa = 8.24TQDD367 pKa = 3.49INTSLALSSFDD378 pKa = 3.34HH379 pKa = 5.54VTFTRR384 pKa = 11.84VDD386 pKa = 3.82CPTDD390 pKa = 2.89IAAFDD395 pKa = 3.68KK396 pKa = 11.34GLVIDD401 pKa = 4.47SGLGASLFITIYY413 pKa = 10.57KK414 pKa = 10.14RR415 pKa = 11.84LNLVTKK421 pKa = 7.94MLRR424 pKa = 11.84LVEE427 pKa = 4.12EE428 pKa = 4.46LNEE431 pKa = 4.11YY432 pKa = 10.02TYY434 pKa = 11.01TPADD438 pKa = 3.71FVQQ441 pKa = 3.78
MM1 pKa = 7.16TRR3 pKa = 11.84FIILSCVLCIAISSVATTMAPNVSGNDD30 pKa = 3.3MYY32 pKa = 11.29FEE34 pKa = 4.5NSGSGSSEE42 pKa = 3.74YY43 pKa = 10.74DD44 pKa = 4.04DD45 pKa = 5.13EE46 pKa = 5.39DD47 pKa = 4.35LSSGEE52 pKa = 4.14SDD54 pKa = 3.4MNSTILTEE62 pKa = 4.18NMEE65 pKa = 4.5STSNASDD72 pKa = 3.34GTDD75 pKa = 3.01EE76 pKa = 5.08YY77 pKa = 10.09NTQVTTFPNDD87 pKa = 2.76VTTDD91 pKa = 3.95FTLTPNNNYY100 pKa = 8.14STLSDD105 pKa = 3.34NRR107 pKa = 11.84SPEE110 pKa = 4.28TKK112 pKa = 10.58NPTDD116 pKa = 3.68TQTSTTPTEE125 pKa = 3.84TSISIKK131 pKa = 10.14TPTQTSAQMIQLTNTPTPTEE151 pKa = 4.1LPNTEE156 pKa = 4.38SPSLTTEE163 pKa = 3.97AVTSMKK169 pKa = 10.27TYY171 pKa = 10.29ILTSRR176 pKa = 11.84EE177 pKa = 4.09SSSKK181 pKa = 10.17SSEE184 pKa = 4.23DD185 pKa = 3.06IKK187 pKa = 11.3SEE189 pKa = 4.28NITSTSNTVYY199 pKa = 9.21KK200 pKa = 10.71TSDD203 pKa = 3.17TDD205 pKa = 3.43VFTYY209 pKa = 8.76QTTNASPTTISTEE222 pKa = 4.04TSVPTTPDD230 pKa = 3.16TKK232 pKa = 11.14SLDD235 pKa = 3.9SNKK238 pKa = 8.56VTDD241 pKa = 4.39SVTGTNSTMVTNNSSATIHH260 pKa = 6.53ASTPISHH267 pKa = 7.33TSTVVYY273 pKa = 8.7ITPHH277 pKa = 6.73NITLPDD283 pKa = 3.5TSTVSIAPHH292 pKa = 6.78NITSFDD298 pKa = 3.46TSTVSTIPHH307 pKa = 5.49VTPSDD312 pKa = 3.73YY313 pKa = 11.51NHH315 pKa = 7.12NITNHH320 pKa = 5.4NVTLYY325 pKa = 10.82DD326 pKa = 3.61YY327 pKa = 10.49DD328 pKa = 4.07YY329 pKa = 11.68KK330 pKa = 11.29NLQTTSSVSVSDD342 pKa = 4.86LYY344 pKa = 10.84ISTTIQTTPLFTPVPCISNYY364 pKa = 8.24TQDD367 pKa = 3.49INTSLALSSFDD378 pKa = 3.34HH379 pKa = 5.54VTFTRR384 pKa = 11.84VDD386 pKa = 3.82CPTDD390 pKa = 2.89IAAFDD395 pKa = 3.68KK396 pKa = 11.34GLVIDD401 pKa = 4.47SGLGASLFITIYY413 pKa = 10.57KK414 pKa = 10.14RR415 pKa = 11.84LNLVTKK421 pKa = 7.94MLRR424 pKa = 11.84LVEE427 pKa = 4.12EE428 pKa = 4.46LNEE431 pKa = 4.11YY432 pKa = 10.02TYY434 pKa = 11.01TPADD438 pKa = 3.71FVQQ441 pKa = 3.78
Molecular weight: 47.84 kDa
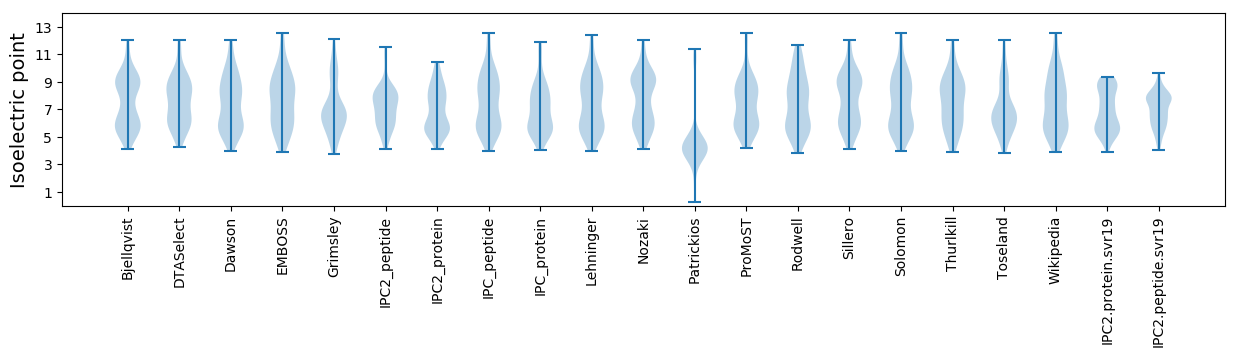
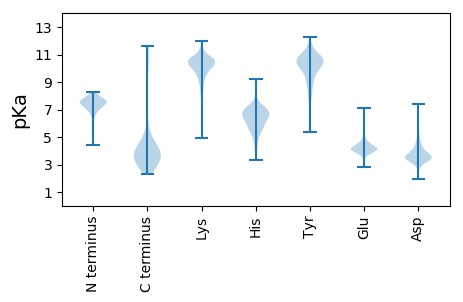
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7YNI0|K7YNI0_RCMVE E31 OS=Rat cytomegalovirus (isolate England) OX=1261657 GN=E31 PE=3 SV=1
MM1 pKa = 7.54TSMCVRR7 pKa = 11.84RR8 pKa = 11.84EE9 pKa = 3.55LFAPHH14 pKa = 6.9RR15 pKa = 11.84SGVEE19 pKa = 3.56NHH21 pKa = 5.91PRR23 pKa = 11.84AMNGKK28 pKa = 9.58GVDD31 pKa = 3.52NPNRR35 pKa = 11.84SLPVSRR41 pKa = 11.84RR42 pKa = 11.84SRR44 pKa = 11.84AMGKK48 pKa = 8.66MGEE51 pKa = 4.5TTGSRR56 pKa = 11.84MRR58 pKa = 11.84VSTVFALACVVVIAVTALPGADD80 pKa = 2.79AVVDD84 pKa = 3.92AWSTGCEE91 pKa = 3.97PARR94 pKa = 11.84IGQMCKK100 pKa = 8.82MYY102 pKa = 11.02ARR104 pKa = 11.84ASRR107 pKa = 11.84GNIPSPPPQYY117 pKa = 10.81VSWSRR122 pKa = 11.84IGASSRR128 pKa = 11.84NEE130 pKa = 3.63VLLLSRR136 pKa = 11.84TEE138 pKa = 4.23VPWLGPPIKK147 pKa = 9.69TRR149 pKa = 11.84LPRR152 pKa = 11.84KK153 pKa = 9.37SGMRR157 pKa = 11.84GITGTAVSSKK167 pKa = 10.84GVVTSALIFNVTKK180 pKa = 10.04NTDD183 pKa = 1.92GWYY186 pKa = 9.63RR187 pKa = 11.84CNYY190 pKa = 9.2YY191 pKa = 10.72LDD193 pKa = 4.03GSVAEE198 pKa = 3.92FHH200 pKa = 7.38VNITAAATEE209 pKa = 4.32GGKK212 pKa = 9.95
MM1 pKa = 7.54TSMCVRR7 pKa = 11.84RR8 pKa = 11.84EE9 pKa = 3.55LFAPHH14 pKa = 6.9RR15 pKa = 11.84SGVEE19 pKa = 3.56NHH21 pKa = 5.91PRR23 pKa = 11.84AMNGKK28 pKa = 9.58GVDD31 pKa = 3.52NPNRR35 pKa = 11.84SLPVSRR41 pKa = 11.84RR42 pKa = 11.84SRR44 pKa = 11.84AMGKK48 pKa = 8.66MGEE51 pKa = 4.5TTGSRR56 pKa = 11.84MRR58 pKa = 11.84VSTVFALACVVVIAVTALPGADD80 pKa = 2.79AVVDD84 pKa = 3.92AWSTGCEE91 pKa = 3.97PARR94 pKa = 11.84IGQMCKK100 pKa = 8.82MYY102 pKa = 11.02ARR104 pKa = 11.84ASRR107 pKa = 11.84GNIPSPPPQYY117 pKa = 10.81VSWSRR122 pKa = 11.84IGASSRR128 pKa = 11.84NEE130 pKa = 3.63VLLLSRR136 pKa = 11.84TEE138 pKa = 4.23VPWLGPPIKK147 pKa = 9.69TRR149 pKa = 11.84LPRR152 pKa = 11.84KK153 pKa = 9.37SGMRR157 pKa = 11.84GITGTAVSSKK167 pKa = 10.84GVVTSALIFNVTKK180 pKa = 10.04NTDD183 pKa = 1.92GWYY186 pKa = 9.63RR187 pKa = 11.84CNYY190 pKa = 9.2YY191 pKa = 10.72LDD193 pKa = 4.03GSVAEE198 pKa = 3.92FHH200 pKa = 7.38VNITAAATEE209 pKa = 4.32GGKK212 pKa = 9.95
Molecular weight: 22.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
55952 |
55 |
2029 |
408.4 |
45.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.962 ± 0.202 | 2.418 ± 0.107 |
6.075 ± 0.141 | 5.392 ± 0.135 |
4.339 ± 0.138 | 5.601 ± 0.268 |
2.407 ± 0.089 | 6.061 ± 0.194 |
4.624 ± 0.14 | 8.632 ± 0.201 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.42 ± 0.08 | 4.438 ± 0.124 |
4.86 ± 0.157 | 2.731 ± 0.121 |
6.908 ± 0.185 | 8.377 ± 0.212 |
6.836 ± 0.256 | 7.326 ± 0.181 |
1.013 ± 0.07 | 3.58 ± 0.111 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
