
Marinactinospora thermotolerans DSM 45154
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Marinactinospora; Marinactinospora thermotolerans
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
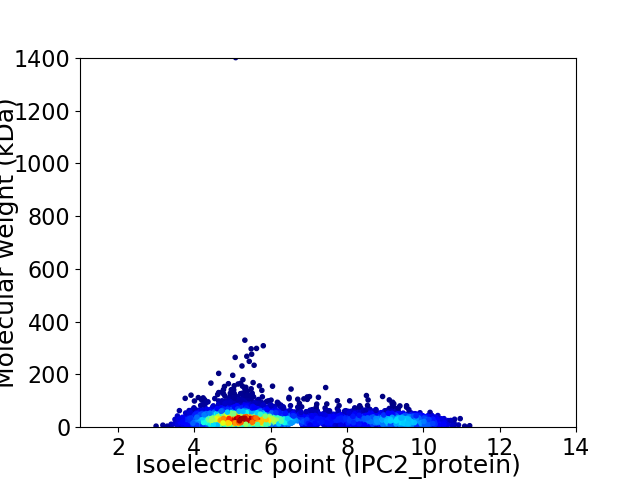
Virtual 2D-PAGE plot for 4956 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
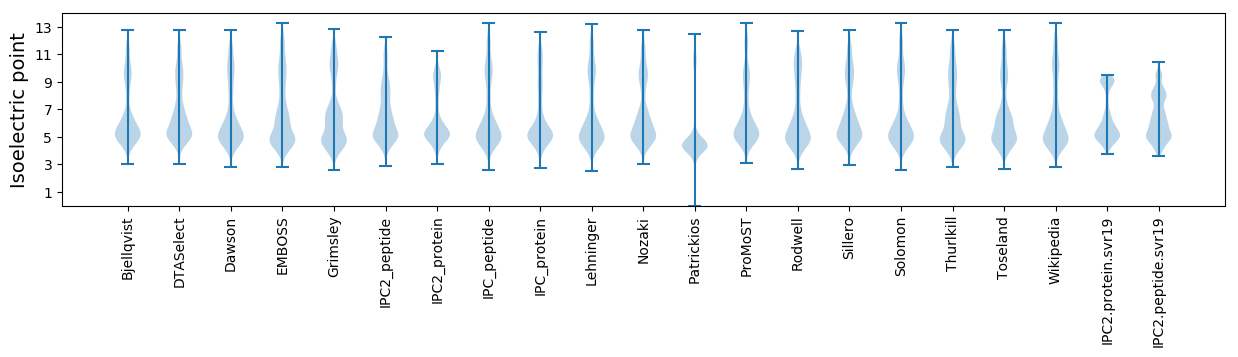
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1T4RVP6|A0A1T4RVP6_9ACTN Uncharacterized protein OS=Marinactinospora thermotolerans DSM 45154 OX=1122192 GN=SAMN02745673_03036 PE=4 SV=1
MM1 pKa = 6.54TTPARR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84ATIAASLPLLLLAGGCSYY26 pKa = 11.38LSGEE30 pKa = 4.43SQGADD35 pKa = 3.61PQSADD40 pKa = 3.48CDD42 pKa = 4.84AYY44 pKa = 10.31EE45 pKa = 3.79QWQGHH50 pKa = 7.05DD51 pKa = 3.46GTTVSIYY58 pKa = 11.11ASIRR62 pKa = 11.84DD63 pKa = 3.67QEE65 pKa = 4.58AEE67 pKa = 4.05LLEE70 pKa = 4.42EE71 pKa = 4.25SWSEE75 pKa = 3.82FSSCTGIDD83 pKa = 2.89IAYY86 pKa = 8.16EE87 pKa = 3.97GSGEE91 pKa = 4.14FEE93 pKa = 3.86AQIQVKK99 pKa = 9.99VDD101 pKa = 3.32GGNAPDD107 pKa = 4.45LAFFPQPGLLARR119 pKa = 11.84FAQSGDD125 pKa = 3.6AIALPEE131 pKa = 4.21SVRR134 pKa = 11.84ANAEE138 pKa = 4.4SGWSEE143 pKa = 3.81DD144 pKa = 2.95WLNYY148 pKa = 8.62ATVDD152 pKa = 3.45GEE154 pKa = 5.15LYY156 pKa = 8.28GTPLGANVKK165 pKa = 10.15SFVWYY170 pKa = 10.57SPGLFSDD177 pKa = 3.81NGYY180 pKa = 8.19ATPTTWDD187 pKa = 3.54EE188 pKa = 4.48LIEE191 pKa = 4.78ISDD194 pKa = 3.67AMVEE198 pKa = 4.25DD199 pKa = 6.26GIKK202 pKa = 9.4PWCAGIEE209 pKa = 4.21SGDD212 pKa = 3.58ATGWPATDD220 pKa = 2.91WVEE223 pKa = 4.35NIMLRR228 pKa = 11.84EE229 pKa = 4.37HH230 pKa = 6.51GPEE233 pKa = 5.41VYY235 pKa = 10.23DD236 pKa = 2.98QWVDD240 pKa = 3.28HH241 pKa = 6.96EE242 pKa = 5.2IPFDD246 pKa = 4.21DD247 pKa = 4.45PAVAEE252 pKa = 4.18ALDD255 pKa = 3.96RR256 pKa = 11.84VDD258 pKa = 5.22SILRR262 pKa = 11.84NPDD265 pKa = 3.36YY266 pKa = 11.7VNGGHH271 pKa = 7.0GEE273 pKa = 4.19VQSIATTSFQDD284 pKa = 3.0AGTPILDD291 pKa = 4.19RR292 pKa = 11.84QCGMYY297 pKa = 11.08LMGSFYY303 pKa = 10.81AAQWPEE309 pKa = 3.75GTTVAEE315 pKa = 4.95DD316 pKa = 3.73GDD318 pKa = 4.3VYY320 pKa = 11.42AFNLPPIQEE329 pKa = 4.13EE330 pKa = 4.02HH331 pKa = 5.8GTPVLGGGEE340 pKa = 4.33FIGAFADD347 pKa = 4.07RR348 pKa = 11.84PEE350 pKa = 4.16VVAVQEE356 pKa = 4.08YY357 pKa = 10.58LSTVEE362 pKa = 4.15YY363 pKa = 10.63ANSRR367 pKa = 11.84ASLGSWFSAHH377 pKa = 7.35RR378 pKa = 11.84DD379 pKa = 3.28LDD381 pKa = 4.21LDD383 pKa = 4.1LLEE386 pKa = 5.47APTDD390 pKa = 3.58RR391 pKa = 11.84LGAEE395 pKa = 4.18TLRR398 pKa = 11.84DD399 pKa = 3.41SEE401 pKa = 4.46AVFRR405 pKa = 11.84FDD407 pKa = 4.91GSDD410 pKa = 3.28MMPASVGAGTFWRR423 pKa = 11.84GMTNWINGDD432 pKa = 3.51EE433 pKa = 4.38TEE435 pKa = 4.19EE436 pKa = 3.74TLAYY440 pKa = 9.59IEE442 pKa = 4.65EE443 pKa = 4.46SWPSSS448 pKa = 3.23
MM1 pKa = 6.54TTPARR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84ATIAASLPLLLLAGGCSYY26 pKa = 11.38LSGEE30 pKa = 4.43SQGADD35 pKa = 3.61PQSADD40 pKa = 3.48CDD42 pKa = 4.84AYY44 pKa = 10.31EE45 pKa = 3.79QWQGHH50 pKa = 7.05DD51 pKa = 3.46GTTVSIYY58 pKa = 11.11ASIRR62 pKa = 11.84DD63 pKa = 3.67QEE65 pKa = 4.58AEE67 pKa = 4.05LLEE70 pKa = 4.42EE71 pKa = 4.25SWSEE75 pKa = 3.82FSSCTGIDD83 pKa = 2.89IAYY86 pKa = 8.16EE87 pKa = 3.97GSGEE91 pKa = 4.14FEE93 pKa = 3.86AQIQVKK99 pKa = 9.99VDD101 pKa = 3.32GGNAPDD107 pKa = 4.45LAFFPQPGLLARR119 pKa = 11.84FAQSGDD125 pKa = 3.6AIALPEE131 pKa = 4.21SVRR134 pKa = 11.84ANAEE138 pKa = 4.4SGWSEE143 pKa = 3.81DD144 pKa = 2.95WLNYY148 pKa = 8.62ATVDD152 pKa = 3.45GEE154 pKa = 5.15LYY156 pKa = 8.28GTPLGANVKK165 pKa = 10.15SFVWYY170 pKa = 10.57SPGLFSDD177 pKa = 3.81NGYY180 pKa = 8.19ATPTTWDD187 pKa = 3.54EE188 pKa = 4.48LIEE191 pKa = 4.78ISDD194 pKa = 3.67AMVEE198 pKa = 4.25DD199 pKa = 6.26GIKK202 pKa = 9.4PWCAGIEE209 pKa = 4.21SGDD212 pKa = 3.58ATGWPATDD220 pKa = 2.91WVEE223 pKa = 4.35NIMLRR228 pKa = 11.84EE229 pKa = 4.37HH230 pKa = 6.51GPEE233 pKa = 5.41VYY235 pKa = 10.23DD236 pKa = 2.98QWVDD240 pKa = 3.28HH241 pKa = 6.96EE242 pKa = 5.2IPFDD246 pKa = 4.21DD247 pKa = 4.45PAVAEE252 pKa = 4.18ALDD255 pKa = 3.96RR256 pKa = 11.84VDD258 pKa = 5.22SILRR262 pKa = 11.84NPDD265 pKa = 3.36YY266 pKa = 11.7VNGGHH271 pKa = 7.0GEE273 pKa = 4.19VQSIATTSFQDD284 pKa = 3.0AGTPILDD291 pKa = 4.19RR292 pKa = 11.84QCGMYY297 pKa = 11.08LMGSFYY303 pKa = 10.81AAQWPEE309 pKa = 3.75GTTVAEE315 pKa = 4.95DD316 pKa = 3.73GDD318 pKa = 4.3VYY320 pKa = 11.42AFNLPPIQEE329 pKa = 4.13EE330 pKa = 4.02HH331 pKa = 5.8GTPVLGGGEE340 pKa = 4.33FIGAFADD347 pKa = 4.07RR348 pKa = 11.84PEE350 pKa = 4.16VVAVQEE356 pKa = 4.08YY357 pKa = 10.58LSTVEE362 pKa = 4.15YY363 pKa = 10.63ANSRR367 pKa = 11.84ASLGSWFSAHH377 pKa = 7.35RR378 pKa = 11.84DD379 pKa = 3.28LDD381 pKa = 4.21LDD383 pKa = 4.1LLEE386 pKa = 5.47APTDD390 pKa = 3.58RR391 pKa = 11.84LGAEE395 pKa = 4.18TLRR398 pKa = 11.84DD399 pKa = 3.41SEE401 pKa = 4.46AVFRR405 pKa = 11.84FDD407 pKa = 4.91GSDD410 pKa = 3.28MMPASVGAGTFWRR423 pKa = 11.84GMTNWINGDD432 pKa = 3.51EE433 pKa = 4.38TEE435 pKa = 4.19EE436 pKa = 3.74TLAYY440 pKa = 9.59IEE442 pKa = 4.65EE443 pKa = 4.46SWPSSS448 pKa = 3.23
Molecular weight: 48.68 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1T4N2Y3|A0A1T4N2Y3_9ACTN Glutaredoxin OS=Marinactinospora thermotolerans DSM 45154 OX=1122192 GN=SAMN02745673_01245 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.55LLKK22 pKa = 7.77KK23 pKa = 9.13TRR25 pKa = 11.84IARR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.8KK32 pKa = 9.86
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.55LLKK22 pKa = 7.77KK23 pKa = 9.13TRR25 pKa = 11.84IARR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.8KK32 pKa = 9.86
Molecular weight: 3.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1596438 |
29 |
12976 |
322.1 |
34.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.249 ± 0.05 | 0.75 ± 0.011 |
5.993 ± 0.036 | 6.375 ± 0.037 |
2.737 ± 0.021 | 9.527 ± 0.034 |
2.281 ± 0.015 | 3.46 ± 0.026 |
1.515 ± 0.021 | 10.442 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.776 ± 0.015 | 1.628 ± 0.017 |
6.279 ± 0.034 | 2.404 ± 0.023 |
8.803 ± 0.042 | 4.96 ± 0.027 |
5.661 ± 0.031 | 8.746 ± 0.041 |
1.462 ± 0.016 | 1.951 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
