
Beihai tombus-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
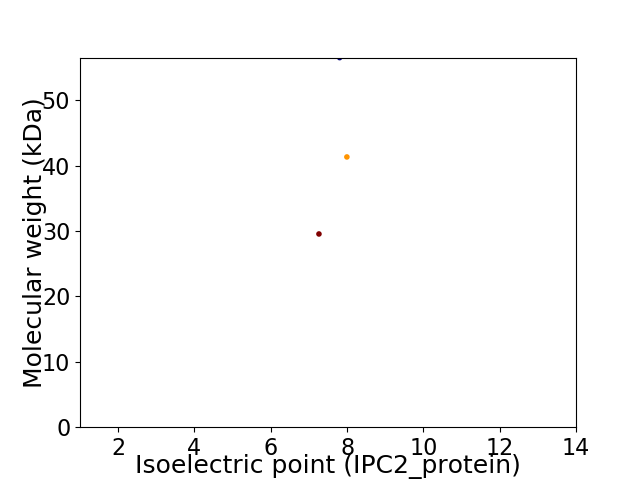
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG87|A0A1L3KG87_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 1 OX=1922712 PE=4 SV=1
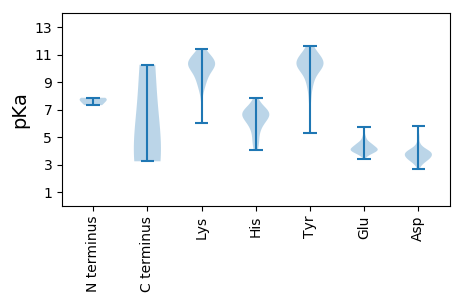
MM1 pKa = 7.68SRR3 pKa = 11.84GTGFACLLATSEE15 pKa = 4.43CIKK18 pKa = 10.7LDD20 pKa = 3.4VLHH23 pKa = 6.68GVGHH27 pKa = 7.11ISPSLLLDD35 pKa = 3.89LPVEE39 pKa = 4.12SSTIIEE45 pKa = 4.47SVHH48 pKa = 6.79RR49 pKa = 11.84EE50 pKa = 3.85PNYY53 pKa = 11.2AKK55 pKa = 10.6YY56 pKa = 10.62GWFAVGGLAAYY67 pKa = 9.81AGYY70 pKa = 10.23RR71 pKa = 11.84YY72 pKa = 10.0LRR74 pKa = 11.84NRR76 pKa = 11.84YY77 pKa = 7.27SQRR80 pKa = 11.84LNHH83 pKa = 5.27TQYY86 pKa = 11.61DD87 pKa = 3.88KK88 pKa = 11.41LHH90 pKa = 6.62EE91 pKa = 4.36EE92 pKa = 4.21VATLLEE98 pKa = 4.61VNDD101 pKa = 4.27EE102 pKa = 4.27PVDD105 pKa = 3.83PEE107 pKa = 4.99DD108 pKa = 3.99PVEE111 pKa = 4.22TTNEE115 pKa = 4.13DD116 pKa = 3.19GSVEE120 pKa = 4.31VVVRR124 pKa = 11.84RR125 pKa = 11.84PRR127 pKa = 11.84KK128 pKa = 8.48RR129 pKa = 11.84SHH131 pKa = 6.36RR132 pKa = 11.84KK133 pKa = 8.57HH134 pKa = 6.68RR135 pKa = 11.84FITDD139 pKa = 4.52CIHH142 pKa = 4.76MTKK145 pKa = 10.35AHH147 pKa = 6.79FGGCPKK153 pKa = 9.6FTEE156 pKa = 4.49SNQIAVSRR164 pKa = 11.84HH165 pKa = 4.06VYY167 pKa = 9.04EE168 pKa = 3.93LCKK171 pKa = 10.11EE172 pKa = 4.5RR173 pKa = 11.84KK174 pKa = 9.12CLPHH178 pKa = 4.33QTRR181 pKa = 11.84TIMAVAVPAVFSPDD195 pKa = 3.69DD196 pKa = 4.39LDD198 pKa = 3.79VSSAQAYY205 pKa = 9.98NIGLRR210 pKa = 11.84RR211 pKa = 11.84EE212 pKa = 3.98RR213 pKa = 11.84RR214 pKa = 11.84SMVQKK219 pKa = 10.96AMEE222 pKa = 4.47GPRR225 pKa = 11.84WYY227 pKa = 10.06EE228 pKa = 3.71GLLDD232 pKa = 3.94NPLNAVAWRR241 pKa = 11.84EE242 pKa = 3.87AMDD245 pKa = 3.67WLFGLPSWKK254 pKa = 10.28AIRR257 pKa = 11.84AVKK260 pKa = 10.26
MM1 pKa = 7.68SRR3 pKa = 11.84GTGFACLLATSEE15 pKa = 4.43CIKK18 pKa = 10.7LDD20 pKa = 3.4VLHH23 pKa = 6.68GVGHH27 pKa = 7.11ISPSLLLDD35 pKa = 3.89LPVEE39 pKa = 4.12SSTIIEE45 pKa = 4.47SVHH48 pKa = 6.79RR49 pKa = 11.84EE50 pKa = 3.85PNYY53 pKa = 11.2AKK55 pKa = 10.6YY56 pKa = 10.62GWFAVGGLAAYY67 pKa = 9.81AGYY70 pKa = 10.23RR71 pKa = 11.84YY72 pKa = 10.0LRR74 pKa = 11.84NRR76 pKa = 11.84YY77 pKa = 7.27SQRR80 pKa = 11.84LNHH83 pKa = 5.27TQYY86 pKa = 11.61DD87 pKa = 3.88KK88 pKa = 11.41LHH90 pKa = 6.62EE91 pKa = 4.36EE92 pKa = 4.21VATLLEE98 pKa = 4.61VNDD101 pKa = 4.27EE102 pKa = 4.27PVDD105 pKa = 3.83PEE107 pKa = 4.99DD108 pKa = 3.99PVEE111 pKa = 4.22TTNEE115 pKa = 4.13DD116 pKa = 3.19GSVEE120 pKa = 4.31VVVRR124 pKa = 11.84RR125 pKa = 11.84PRR127 pKa = 11.84KK128 pKa = 8.48RR129 pKa = 11.84SHH131 pKa = 6.36RR132 pKa = 11.84KK133 pKa = 8.57HH134 pKa = 6.68RR135 pKa = 11.84FITDD139 pKa = 4.52CIHH142 pKa = 4.76MTKK145 pKa = 10.35AHH147 pKa = 6.79FGGCPKK153 pKa = 9.6FTEE156 pKa = 4.49SNQIAVSRR164 pKa = 11.84HH165 pKa = 4.06VYY167 pKa = 9.04EE168 pKa = 3.93LCKK171 pKa = 10.11EE172 pKa = 4.5RR173 pKa = 11.84KK174 pKa = 9.12CLPHH178 pKa = 4.33QTRR181 pKa = 11.84TIMAVAVPAVFSPDD195 pKa = 3.69DD196 pKa = 4.39LDD198 pKa = 3.79VSSAQAYY205 pKa = 9.98NIGLRR210 pKa = 11.84RR211 pKa = 11.84EE212 pKa = 3.98RR213 pKa = 11.84RR214 pKa = 11.84SMVQKK219 pKa = 10.96AMEE222 pKa = 4.47GPRR225 pKa = 11.84WYY227 pKa = 10.06EE228 pKa = 3.71GLLDD232 pKa = 3.94NPLNAVAWRR241 pKa = 11.84EE242 pKa = 3.87AMDD245 pKa = 3.67WLFGLPSWKK254 pKa = 10.28AIRR257 pKa = 11.84AVKK260 pKa = 10.26
Molecular weight: 29.56 kDa
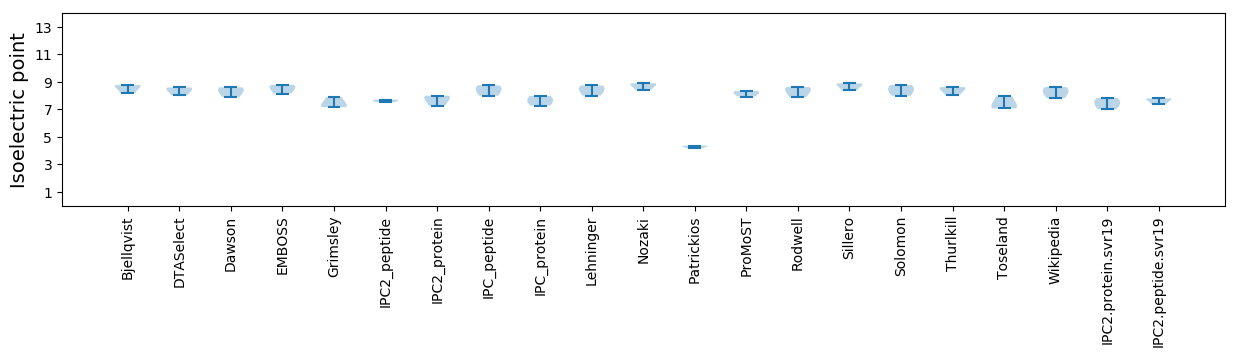
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG87|A0A1L3KG87_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 1 OX=1922712 PE=4 SV=1
MM1 pKa = 7.86AIVPYY6 pKa = 10.35NAAGQLASLITHH18 pKa = 7.16PGIQQKK24 pKa = 7.78VAKK27 pKa = 9.29FASQGLPWASKK38 pKa = 10.23LLSAGWQKK46 pKa = 9.24LTKK49 pKa = 9.82GQRR52 pKa = 11.84KK53 pKa = 6.0QARR56 pKa = 11.84NALNLQPKK64 pKa = 8.51IQANMVPVEE73 pKa = 4.04VGISKK78 pKa = 10.33SAIVKK83 pKa = 9.89GRR85 pKa = 11.84IPRR88 pKa = 11.84YY89 pKa = 8.33KK90 pKa = 10.01QSAGYY95 pKa = 7.96VTISHH100 pKa = 6.7RR101 pKa = 11.84EE102 pKa = 3.84YY103 pKa = 10.44LAEE106 pKa = 4.6AEE108 pKa = 4.69LVNGTDD114 pKa = 2.7IRR116 pKa = 11.84GYY118 pKa = 10.13VLNPSNPRR126 pKa = 11.84AFTWLCTFARR136 pKa = 11.84NFEE139 pKa = 4.23KK140 pKa = 11.0YY141 pKa = 8.89RR142 pKa = 11.84FRR144 pKa = 11.84NIRR147 pKa = 11.84LTYY150 pKa = 9.91VPQAPTTAAGRR161 pKa = 11.84VILAYY166 pKa = 10.47DD167 pKa = 3.91KK168 pKa = 11.18DD169 pKa = 4.01STDD172 pKa = 3.39ILPSGKK178 pKa = 10.19AEE180 pKa = 4.11LYY182 pKa = 10.23SYY184 pKa = 10.63QGTVDD189 pKa = 3.51TPVWQGTSIAFPCDD203 pKa = 2.9NTLRR207 pKa = 11.84FVDD210 pKa = 3.86SSNTAEE216 pKa = 4.12TRR218 pKa = 11.84LVDD221 pKa = 4.15LGQFITCTSAVSAFNGGEE239 pKa = 3.79FFIDD243 pKa = 3.63YY244 pKa = 9.09TVEE247 pKa = 3.71LHH249 pKa = 6.44TPQQPQPMSEE259 pKa = 4.14RR260 pKa = 11.84IYY262 pKa = 10.01TVVGIPAEE270 pKa = 4.08YY271 pKa = 10.19HH272 pKa = 5.74GDD274 pKa = 3.59LMVVEE279 pKa = 5.26SNNSTTEE286 pKa = 4.71FGIEE290 pKa = 3.8WQSVGFYY297 pKa = 10.19HH298 pKa = 7.54VSITINGDD306 pKa = 3.57CSSMATNTHH315 pKa = 6.84DD316 pKa = 3.82DD317 pKa = 3.21AHH319 pKa = 6.49IVRR322 pKa = 11.84TTSSQNGTTSMTIAMFVRR340 pKa = 11.84VVSFDD345 pKa = 3.23PTAPPGILFTLLSGRR360 pKa = 11.84GSVHH364 pKa = 6.14TYY366 pKa = 5.29VTRR369 pKa = 11.84VSSSVAFSS377 pKa = 3.24
MM1 pKa = 7.86AIVPYY6 pKa = 10.35NAAGQLASLITHH18 pKa = 7.16PGIQQKK24 pKa = 7.78VAKK27 pKa = 9.29FASQGLPWASKK38 pKa = 10.23LLSAGWQKK46 pKa = 9.24LTKK49 pKa = 9.82GQRR52 pKa = 11.84KK53 pKa = 6.0QARR56 pKa = 11.84NALNLQPKK64 pKa = 8.51IQANMVPVEE73 pKa = 4.04VGISKK78 pKa = 10.33SAIVKK83 pKa = 9.89GRR85 pKa = 11.84IPRR88 pKa = 11.84YY89 pKa = 8.33KK90 pKa = 10.01QSAGYY95 pKa = 7.96VTISHH100 pKa = 6.7RR101 pKa = 11.84EE102 pKa = 3.84YY103 pKa = 10.44LAEE106 pKa = 4.6AEE108 pKa = 4.69LVNGTDD114 pKa = 2.7IRR116 pKa = 11.84GYY118 pKa = 10.13VLNPSNPRR126 pKa = 11.84AFTWLCTFARR136 pKa = 11.84NFEE139 pKa = 4.23KK140 pKa = 11.0YY141 pKa = 8.89RR142 pKa = 11.84FRR144 pKa = 11.84NIRR147 pKa = 11.84LTYY150 pKa = 9.91VPQAPTTAAGRR161 pKa = 11.84VILAYY166 pKa = 10.47DD167 pKa = 3.91KK168 pKa = 11.18DD169 pKa = 4.01STDD172 pKa = 3.39ILPSGKK178 pKa = 10.19AEE180 pKa = 4.11LYY182 pKa = 10.23SYY184 pKa = 10.63QGTVDD189 pKa = 3.51TPVWQGTSIAFPCDD203 pKa = 2.9NTLRR207 pKa = 11.84FVDD210 pKa = 3.86SSNTAEE216 pKa = 4.12TRR218 pKa = 11.84LVDD221 pKa = 4.15LGQFITCTSAVSAFNGGEE239 pKa = 3.79FFIDD243 pKa = 3.63YY244 pKa = 9.09TVEE247 pKa = 3.71LHH249 pKa = 6.44TPQQPQPMSEE259 pKa = 4.14RR260 pKa = 11.84IYY262 pKa = 10.01TVVGIPAEE270 pKa = 4.08YY271 pKa = 10.19HH272 pKa = 5.74GDD274 pKa = 3.59LMVVEE279 pKa = 5.26SNNSTTEE286 pKa = 4.71FGIEE290 pKa = 3.8WQSVGFYY297 pKa = 10.19HH298 pKa = 7.54VSITINGDD306 pKa = 3.57CSSMATNTHH315 pKa = 6.84DD316 pKa = 3.82DD317 pKa = 3.21AHH319 pKa = 6.49IVRR322 pKa = 11.84TTSSQNGTTSMTIAMFVRR340 pKa = 11.84VVSFDD345 pKa = 3.23PTAPPGILFTLLSGRR360 pKa = 11.84GSVHH364 pKa = 6.14TYY366 pKa = 5.29VTRR369 pKa = 11.84VSSSVAFSS377 pKa = 3.24
Molecular weight: 41.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1142 |
260 |
505 |
380.7 |
42.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.968 ± 0.254 | 1.839 ± 0.326 |
4.553 ± 0.35 | 5.342 ± 0.797 |
4.203 ± 0.509 | 7.093 ± 0.522 |
2.715 ± 0.507 | 4.904 ± 0.526 |
4.729 ± 0.6 | 7.618 ± 0.566 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.277 ± 0.187 | 4.378 ± 0.314 |
5.517 ± 0.123 | 3.678 ± 0.621 |
6.305 ± 0.764 | 7.618 ± 0.6 |
5.342 ± 1.705 | 8.406 ± 0.078 |
1.751 ± 0.178 | 3.765 ± 0.112 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
