
Epstein-Barr virus (strain AG876) (HHV-4) (Human herpesvirus 4)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; Lymphocryptovirus; Human gammaherpesvirus 4
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
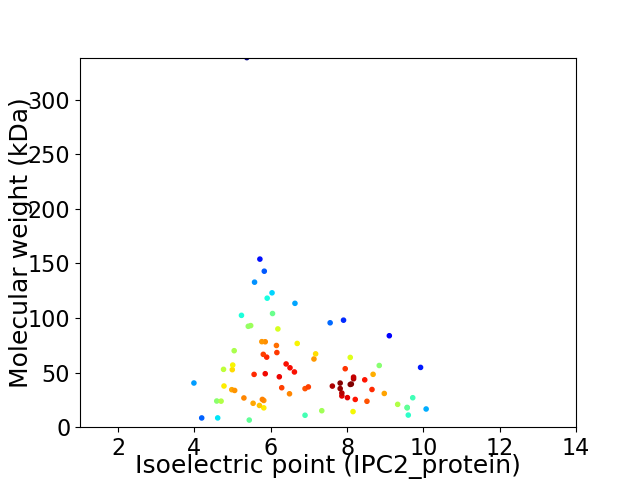
Virtual 2D-PAGE plot for 82 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
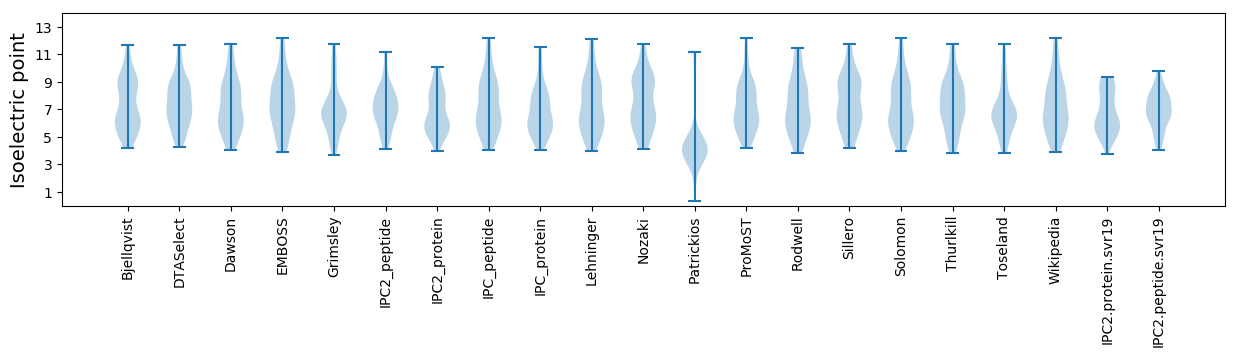
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q1HVB5|BNL2B_EBVA8 Uncharacterized protein BNLF2b OS=Epstein-Barr virus (strain AG876) OX=82830 GN=BNLF2b PE=3 SV=1
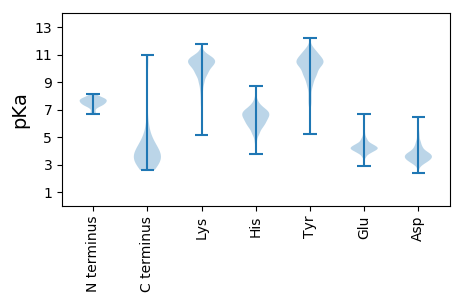
MM1 pKa = 7.25EE2 pKa = 6.06HH3 pKa = 7.2DD4 pKa = 4.84LEE6 pKa = 5.12RR7 pKa = 11.84GPPGPRR13 pKa = 11.84RR14 pKa = 11.84PPRR17 pKa = 11.84GPPLSSSLGLALLLLLLALLFWLYY41 pKa = 10.78IVMSDD46 pKa = 3.13WTGGALLVLYY56 pKa = 10.28SFALILIIIILIIFIFRR73 pKa = 11.84RR74 pKa = 11.84DD75 pKa = 3.73LLCPLGALCLLLLMITLLLIALWNLHH101 pKa = 5.13GQALYY106 pKa = 10.82LGIVLFIFGCLLVLGLWIYY125 pKa = 10.87LLEE128 pKa = 4.3ILWRR132 pKa = 11.84LGATIWQLLAFFLAFFLDD150 pKa = 5.18LILLIIALYY159 pKa = 9.75LQQNWWTLLVDD170 pKa = 5.12LLWLLLFLAILIWMYY185 pKa = 10.53YY186 pKa = 8.99HH187 pKa = 6.85GQRR190 pKa = 11.84HH191 pKa = 5.39SDD193 pKa = 3.35EE194 pKa = 4.43HH195 pKa = 7.75HH196 pKa = 6.96HH197 pKa = 7.46DD198 pKa = 4.28DD199 pKa = 5.28SLPHH203 pKa = 6.25PQQATDD209 pKa = 3.84DD210 pKa = 4.24SGHH213 pKa = 6.22EE214 pKa = 4.08SDD216 pKa = 4.7SNSNEE221 pKa = 3.73GRR223 pKa = 11.84HH224 pKa = 5.67HH225 pKa = 6.75LLVTGAGDD233 pKa = 4.29GPPLCSQNLGAPGGGPDD250 pKa = 5.41NGPQDD255 pKa = 4.7PDD257 pKa = 3.47NTDD260 pKa = 3.56DD261 pKa = 5.97NGPQDD266 pKa = 5.0PDD268 pKa = 3.47NTDD271 pKa = 3.56DD272 pKa = 5.97NGPQDD277 pKa = 5.0PDD279 pKa = 3.47NTDD282 pKa = 3.56DD283 pKa = 5.97NGPQDD288 pKa = 4.43PDD290 pKa = 3.16NTADD294 pKa = 4.25NGPHH298 pKa = 7.25DD299 pKa = 5.18PLPHH303 pKa = 6.42NPSDD307 pKa = 3.85SAGNDD312 pKa = 3.31GGPPNLTEE320 pKa = 3.9EE321 pKa = 4.49VEE323 pKa = 4.4NKK325 pKa = 10.5GGDD328 pKa = 3.31RR329 pKa = 11.84GPPSMTDD336 pKa = 2.93GGGGDD341 pKa = 3.49PHH343 pKa = 8.36LPTLLLGTSGSGGDD357 pKa = 4.67DD358 pKa = 4.4DD359 pKa = 6.1DD360 pKa = 4.11PHH362 pKa = 8.7GPVQLSYY369 pKa = 11.75YY370 pKa = 10.41DD371 pKa = 3.38
MM1 pKa = 7.25EE2 pKa = 6.06HH3 pKa = 7.2DD4 pKa = 4.84LEE6 pKa = 5.12RR7 pKa = 11.84GPPGPRR13 pKa = 11.84RR14 pKa = 11.84PPRR17 pKa = 11.84GPPLSSSLGLALLLLLLALLFWLYY41 pKa = 10.78IVMSDD46 pKa = 3.13WTGGALLVLYY56 pKa = 10.28SFALILIIIILIIFIFRR73 pKa = 11.84RR74 pKa = 11.84DD75 pKa = 3.73LLCPLGALCLLLLMITLLLIALWNLHH101 pKa = 5.13GQALYY106 pKa = 10.82LGIVLFIFGCLLVLGLWIYY125 pKa = 10.87LLEE128 pKa = 4.3ILWRR132 pKa = 11.84LGATIWQLLAFFLAFFLDD150 pKa = 5.18LILLIIALYY159 pKa = 9.75LQQNWWTLLVDD170 pKa = 5.12LLWLLLFLAILIWMYY185 pKa = 10.53YY186 pKa = 8.99HH187 pKa = 6.85GQRR190 pKa = 11.84HH191 pKa = 5.39SDD193 pKa = 3.35EE194 pKa = 4.43HH195 pKa = 7.75HH196 pKa = 6.96HH197 pKa = 7.46DD198 pKa = 4.28DD199 pKa = 5.28SLPHH203 pKa = 6.25PQQATDD209 pKa = 3.84DD210 pKa = 4.24SGHH213 pKa = 6.22EE214 pKa = 4.08SDD216 pKa = 4.7SNSNEE221 pKa = 3.73GRR223 pKa = 11.84HH224 pKa = 5.67HH225 pKa = 6.75LLVTGAGDD233 pKa = 4.29GPPLCSQNLGAPGGGPDD250 pKa = 5.41NGPQDD255 pKa = 4.7PDD257 pKa = 3.47NTDD260 pKa = 3.56DD261 pKa = 5.97NGPQDD266 pKa = 5.0PDD268 pKa = 3.47NTDD271 pKa = 3.56DD272 pKa = 5.97NGPQDD277 pKa = 5.0PDD279 pKa = 3.47NTDD282 pKa = 3.56DD283 pKa = 5.97NGPQDD288 pKa = 4.43PDD290 pKa = 3.16NTADD294 pKa = 4.25NGPHH298 pKa = 7.25DD299 pKa = 5.18PLPHH303 pKa = 6.42NPSDD307 pKa = 3.85SAGNDD312 pKa = 3.31GGPPNLTEE320 pKa = 3.9EE321 pKa = 4.49VEE323 pKa = 4.4NKK325 pKa = 10.5GGDD328 pKa = 3.31RR329 pKa = 11.84GPPSMTDD336 pKa = 2.93GGGGDD341 pKa = 3.49PHH343 pKa = 8.36LPTLLLGTSGSGGDD357 pKa = 4.67DD358 pKa = 4.4DD359 pKa = 6.1DD360 pKa = 4.11PHH362 pKa = 8.7GPVQLSYY369 pKa = 11.75YY370 pKa = 10.41DD371 pKa = 3.38
Molecular weight: 40.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P7U1E1|A0A1P7U1E1_EBVA8 Packaging protein UL32 OS=Epstein-Barr virus (strain AG876) OX=82830 PE=3 SV=1
MM1 pKa = 7.62ARR3 pKa = 11.84RR4 pKa = 11.84LPKK7 pKa = 9.12PTLQGRR13 pKa = 11.84LEE15 pKa = 4.27ADD17 pKa = 3.98FPDD20 pKa = 4.6SPLLPKK26 pKa = 10.41FQEE29 pKa = 4.16LNQNNLPNDD38 pKa = 3.73VFRR41 pKa = 11.84EE42 pKa = 4.15AQRR45 pKa = 11.84SYY47 pKa = 11.54LVFLTSQFCYY57 pKa = 10.33EE58 pKa = 4.52EE59 pKa = 4.17YY60 pKa = 9.89VQRR63 pKa = 11.84TFGVPRR69 pKa = 11.84RR70 pKa = 11.84QRR72 pKa = 11.84AIDD75 pKa = 3.21KK76 pKa = 9.86RR77 pKa = 11.84QRR79 pKa = 11.84ASVAGAGAHH88 pKa = 5.34AHH90 pKa = 6.49LGGSSATPVQQAQAAASAGTGALASSAPSTAVAQSATPSVSSSISSLRR138 pKa = 11.84AATSGATAAASAAAAVDD155 pKa = 3.72TGSGGGGQPQDD166 pKa = 3.46TAPRR170 pKa = 11.84GARR173 pKa = 11.84KK174 pKa = 9.13KK175 pKa = 8.65QQ176 pKa = 3.08
MM1 pKa = 7.62ARR3 pKa = 11.84RR4 pKa = 11.84LPKK7 pKa = 9.12PTLQGRR13 pKa = 11.84LEE15 pKa = 4.27ADD17 pKa = 3.98FPDD20 pKa = 4.6SPLLPKK26 pKa = 10.41FQEE29 pKa = 4.16LNQNNLPNDD38 pKa = 3.73VFRR41 pKa = 11.84EE42 pKa = 4.15AQRR45 pKa = 11.84SYY47 pKa = 11.54LVFLTSQFCYY57 pKa = 10.33EE58 pKa = 4.52EE59 pKa = 4.17YY60 pKa = 9.89VQRR63 pKa = 11.84TFGVPRR69 pKa = 11.84RR70 pKa = 11.84QRR72 pKa = 11.84AIDD75 pKa = 3.21KK76 pKa = 9.86RR77 pKa = 11.84QRR79 pKa = 11.84ASVAGAGAHH88 pKa = 5.34AHH90 pKa = 6.49LGGSSATPVQQAQAAASAGTGALASSAPSTAVAQSATPSVSSSISSLRR138 pKa = 11.84AATSGATAAASAAAAVDD155 pKa = 3.72TGSGGGGQPQDD166 pKa = 3.46TAPRR170 pKa = 11.84GARR173 pKa = 11.84KK174 pKa = 9.13KK175 pKa = 8.65QQ176 pKa = 3.08
Molecular weight: 18.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
41182 |
60 |
3154 |
502.2 |
54.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.128 ± 0.413 | 1.969 ± 0.156 |
4.507 ± 0.153 | 5.434 ± 0.195 |
3.715 ± 0.168 | 7.418 ± 0.542 |
2.496 ± 0.134 | 3.723 ± 0.155 |
3.132 ± 0.158 | 10.497 ± 0.385 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.998 ± 0.119 | 3.052 ± 0.175 |
7.872 ± 0.433 | 3.943 ± 0.195 |
6.539 ± 0.243 | 7.71 ± 0.215 |
6.318 ± 0.374 | 6.486 ± 0.232 |
1.185 ± 0.078 | 2.875 ± 0.134 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
