
Halonotius sp. J07HN4
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halonotius; unclassified Halonotius
Average proteome isoelectric point is 4.89
Get precalculated fractions of proteins
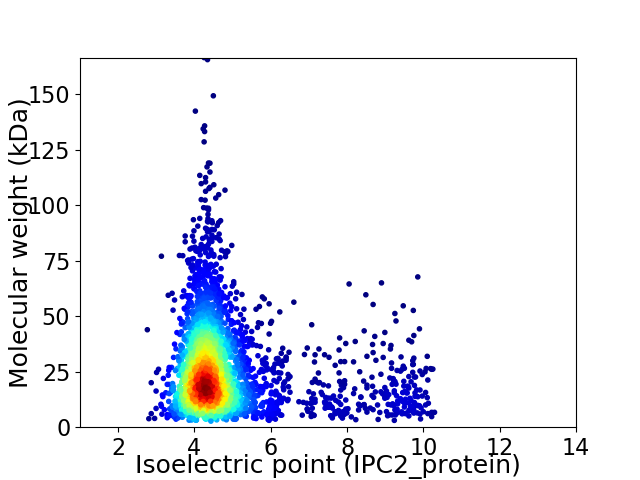
Virtual 2D-PAGE plot for 3224 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U1PA90|U1PA90_9EURY Uncharacterized protein OS=Halonotius sp. J07HN4 OX=1070774 GN=J07HN4v3_02535 PE=4 SV=1
MM1 pKa = 7.79ADD3 pKa = 3.72IDD5 pKa = 4.65LTLDD9 pKa = 3.13WTPNTNHH16 pKa = 6.31TGFYY20 pKa = 9.49VALAEE25 pKa = 5.32GYY27 pKa = 9.85YY28 pKa = 10.33ADD30 pKa = 4.93HH31 pKa = 6.73GLDD34 pKa = 3.62VAIHH38 pKa = 6.08SPAADD43 pKa = 4.03GYY45 pKa = 8.37EE46 pKa = 4.09TTPAKK51 pKa = 10.11QVATGEE57 pKa = 4.21ATLAIAPSEE66 pKa = 4.35SVISYY71 pKa = 7.52QTHH74 pKa = 6.79PDD76 pKa = 3.8YY77 pKa = 11.14PSLTAVAAVCQEE89 pKa = 4.17DD90 pKa = 3.51KK91 pKa = 11.32SAIVTLGDD99 pKa = 3.53SDD101 pKa = 4.83LDD103 pKa = 3.9RR104 pKa = 11.84PADD107 pKa = 3.99LDD109 pKa = 3.66GATYY113 pKa = 10.85ASYY116 pKa = 10.75DD117 pKa = 3.43ARR119 pKa = 11.84FEE121 pKa = 4.14DD122 pKa = 4.36HH123 pKa = 7.18IVRR126 pKa = 11.84QLVRR130 pKa = 11.84NDD132 pKa = 3.7GGDD135 pKa = 3.41GDD137 pKa = 4.43IEE139 pKa = 4.32IVTPPKK145 pKa = 10.41LGIWNTLLDD154 pKa = 4.27GDD156 pKa = 5.6ADD158 pKa = 4.08ATWVFMPWEE167 pKa = 4.49GVLAARR173 pKa = 11.84DD174 pKa = 4.57GIDD177 pKa = 3.42LTPFYY182 pKa = 10.64LDD184 pKa = 3.95TYY186 pKa = 9.83DD187 pKa = 3.37VPYY190 pKa = 10.74GYY192 pKa = 8.78TPLLLARR199 pKa = 11.84PEE201 pKa = 4.29TIADD205 pKa = 3.51GDD207 pKa = 3.98ALADD211 pKa = 4.06FLAATAHH218 pKa = 7.04GYY220 pKa = 9.28QFAVDD225 pKa = 3.8NPDD228 pKa = 3.14RR229 pKa = 11.84AAEE232 pKa = 4.06ILGEE236 pKa = 4.18TAEE239 pKa = 5.36GMDD242 pKa = 4.43NDD244 pKa = 4.93DD245 pKa = 4.01PDD247 pKa = 4.04FLRR250 pKa = 11.84EE251 pKa = 4.17SQHH254 pKa = 6.06EE255 pKa = 4.2LTDD258 pKa = 3.74AYY260 pKa = 10.05LTADD264 pKa = 4.01GQWGRR269 pKa = 11.84MAHH272 pKa = 6.14DD273 pKa = 3.22RR274 pKa = 11.84WDD276 pKa = 3.84AFVDD280 pKa = 3.53WLADD284 pKa = 3.59TDD286 pKa = 3.95ILTTVDD292 pKa = 4.12GDD294 pKa = 4.55PIPADD299 pKa = 3.89EE300 pKa = 5.63LPTDD304 pKa = 3.92DD305 pKa = 6.12LYY307 pKa = 11.41TNEE310 pKa = 4.96LLSDD314 pKa = 3.7SS315 pKa = 4.2
MM1 pKa = 7.79ADD3 pKa = 3.72IDD5 pKa = 4.65LTLDD9 pKa = 3.13WTPNTNHH16 pKa = 6.31TGFYY20 pKa = 9.49VALAEE25 pKa = 5.32GYY27 pKa = 9.85YY28 pKa = 10.33ADD30 pKa = 4.93HH31 pKa = 6.73GLDD34 pKa = 3.62VAIHH38 pKa = 6.08SPAADD43 pKa = 4.03GYY45 pKa = 8.37EE46 pKa = 4.09TTPAKK51 pKa = 10.11QVATGEE57 pKa = 4.21ATLAIAPSEE66 pKa = 4.35SVISYY71 pKa = 7.52QTHH74 pKa = 6.79PDD76 pKa = 3.8YY77 pKa = 11.14PSLTAVAAVCQEE89 pKa = 4.17DD90 pKa = 3.51KK91 pKa = 11.32SAIVTLGDD99 pKa = 3.53SDD101 pKa = 4.83LDD103 pKa = 3.9RR104 pKa = 11.84PADD107 pKa = 3.99LDD109 pKa = 3.66GATYY113 pKa = 10.85ASYY116 pKa = 10.75DD117 pKa = 3.43ARR119 pKa = 11.84FEE121 pKa = 4.14DD122 pKa = 4.36HH123 pKa = 7.18IVRR126 pKa = 11.84QLVRR130 pKa = 11.84NDD132 pKa = 3.7GGDD135 pKa = 3.41GDD137 pKa = 4.43IEE139 pKa = 4.32IVTPPKK145 pKa = 10.41LGIWNTLLDD154 pKa = 4.27GDD156 pKa = 5.6ADD158 pKa = 4.08ATWVFMPWEE167 pKa = 4.49GVLAARR173 pKa = 11.84DD174 pKa = 4.57GIDD177 pKa = 3.42LTPFYY182 pKa = 10.64LDD184 pKa = 3.95TYY186 pKa = 9.83DD187 pKa = 3.37VPYY190 pKa = 10.74GYY192 pKa = 8.78TPLLLARR199 pKa = 11.84PEE201 pKa = 4.29TIADD205 pKa = 3.51GDD207 pKa = 3.98ALADD211 pKa = 4.06FLAATAHH218 pKa = 7.04GYY220 pKa = 9.28QFAVDD225 pKa = 3.8NPDD228 pKa = 3.14RR229 pKa = 11.84AAEE232 pKa = 4.06ILGEE236 pKa = 4.18TAEE239 pKa = 5.36GMDD242 pKa = 4.43NDD244 pKa = 4.93DD245 pKa = 4.01PDD247 pKa = 4.04FLRR250 pKa = 11.84EE251 pKa = 4.17SQHH254 pKa = 6.06EE255 pKa = 4.2LTDD258 pKa = 3.74AYY260 pKa = 10.05LTADD264 pKa = 4.01GQWGRR269 pKa = 11.84MAHH272 pKa = 6.14DD273 pKa = 3.22RR274 pKa = 11.84WDD276 pKa = 3.84AFVDD280 pKa = 3.53WLADD284 pKa = 3.59TDD286 pKa = 3.95ILTTVDD292 pKa = 4.12GDD294 pKa = 4.55PIPADD299 pKa = 3.89EE300 pKa = 5.63LPTDD304 pKa = 3.92DD305 pKa = 6.12LYY307 pKa = 11.41TNEE310 pKa = 4.96LLSDD314 pKa = 3.7SS315 pKa = 4.2
Molecular weight: 34.27 kDa
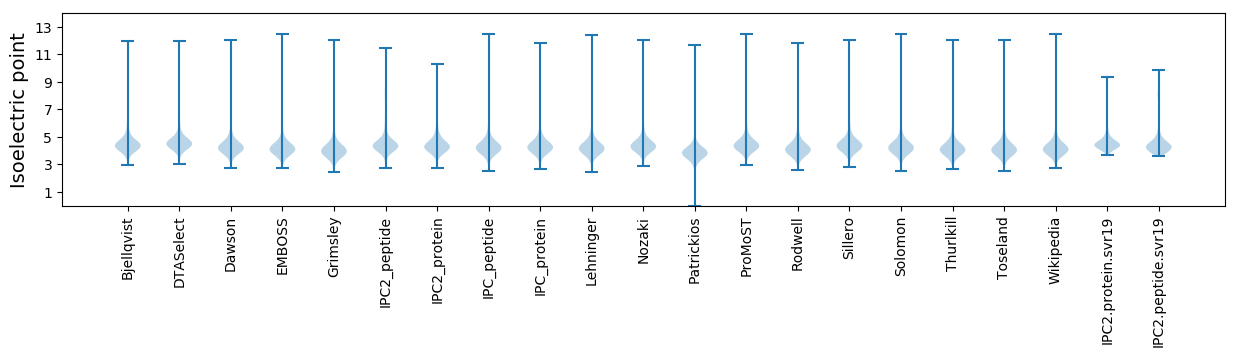
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U1QMW0|U1QMW0_9EURY Metal-dependent carboxypeptidase OS=Halonotius sp. J07HN4 OX=1070774 GN=J07HN4v3_02543 PE=3 SV=1
MM1 pKa = 8.02SLMAAGVVLAAVGGDD16 pKa = 3.61HH17 pKa = 7.51LFGEE21 pKa = 5.03PPNRR25 pKa = 11.84LHH27 pKa = 6.87PVALFGRR34 pKa = 11.84LVGPLDD40 pKa = 4.11RR41 pKa = 11.84SWPAPRR47 pKa = 11.84LIGGVAALSLPLLAAGVVAVVVWIAGRR74 pKa = 11.84SHH76 pKa = 6.78PVVGSVVAAGVLFSTISLRR95 pKa = 11.84LLCATASNVLNQTEE109 pKa = 4.5TNLTDD114 pKa = 3.35ARR116 pKa = 11.84EE117 pKa = 4.12ALLALAGRR125 pKa = 11.84DD126 pKa = 3.72ATKK129 pKa = 10.79LSADD133 pKa = 3.84HH134 pKa = 6.17VRR136 pKa = 11.84SAAVEE141 pKa = 4.13SAAEE145 pKa = 3.97NLSDD149 pKa = 4.35GFVAPLVGFLIASVVGSRR167 pKa = 11.84WLTPGATLAVATAAAAWIKK186 pKa = 10.64AINTMDD192 pKa = 3.37SMVGYY197 pKa = 7.96RR198 pKa = 11.84TKK200 pKa = 10.65PVGWAAARR208 pKa = 11.84LDD210 pKa = 4.26DD211 pKa = 4.6LVMWVPARR219 pKa = 11.84LTAIAISVAALSPDD233 pKa = 3.39PLLVGRR239 pKa = 11.84RR240 pKa = 11.84WARR243 pKa = 11.84EE244 pKa = 3.67PASPNAGWPMATLSAALGVRR264 pKa = 11.84LDD266 pKa = 3.78KK267 pKa = 10.81PGHH270 pKa = 4.84YY271 pKa = 8.94TLNAVASLPTTAAGHH286 pKa = 6.12RR287 pKa = 11.84GVAIVRR293 pKa = 11.84RR294 pKa = 11.84AGWLTALSVAVIAGVV309 pKa = 3.08
MM1 pKa = 8.02SLMAAGVVLAAVGGDD16 pKa = 3.61HH17 pKa = 7.51LFGEE21 pKa = 5.03PPNRR25 pKa = 11.84LHH27 pKa = 6.87PVALFGRR34 pKa = 11.84LVGPLDD40 pKa = 4.11RR41 pKa = 11.84SWPAPRR47 pKa = 11.84LIGGVAALSLPLLAAGVVAVVVWIAGRR74 pKa = 11.84SHH76 pKa = 6.78PVVGSVVAAGVLFSTISLRR95 pKa = 11.84LLCATASNVLNQTEE109 pKa = 4.5TNLTDD114 pKa = 3.35ARR116 pKa = 11.84EE117 pKa = 4.12ALLALAGRR125 pKa = 11.84DD126 pKa = 3.72ATKK129 pKa = 10.79LSADD133 pKa = 3.84HH134 pKa = 6.17VRR136 pKa = 11.84SAAVEE141 pKa = 4.13SAAEE145 pKa = 3.97NLSDD149 pKa = 4.35GFVAPLVGFLIASVVGSRR167 pKa = 11.84WLTPGATLAVATAAAAWIKK186 pKa = 10.64AINTMDD192 pKa = 3.37SMVGYY197 pKa = 7.96RR198 pKa = 11.84TKK200 pKa = 10.65PVGWAAARR208 pKa = 11.84LDD210 pKa = 4.26DD211 pKa = 4.6LVMWVPARR219 pKa = 11.84LTAIAISVAALSPDD233 pKa = 3.39PLLVGRR239 pKa = 11.84RR240 pKa = 11.84WARR243 pKa = 11.84EE244 pKa = 3.67PASPNAGWPMATLSAALGVRR264 pKa = 11.84LDD266 pKa = 3.78KK267 pKa = 10.81PGHH270 pKa = 4.84YY271 pKa = 8.94TLNAVASLPTTAAGHH286 pKa = 6.12RR287 pKa = 11.84GVAIVRR293 pKa = 11.84RR294 pKa = 11.84AGWLTALSVAVIAGVV309 pKa = 3.08
Molecular weight: 31.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
812135 |
25 |
1521 |
251.9 |
27.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.111 ± 0.056 | 0.77 ± 0.015 |
8.628 ± 0.056 | 8.047 ± 0.065 |
3.101 ± 0.029 | 8.223 ± 0.045 |
1.937 ± 0.025 | 4.826 ± 0.036 |
1.923 ± 0.026 | 8.73 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.763 ± 0.019 | 2.533 ± 0.022 |
4.584 ± 0.029 | 2.876 ± 0.026 |
6.043 ± 0.041 | 5.676 ± 0.036 |
7.153 ± 0.038 | 8.379 ± 0.044 |
1.027 ± 0.018 | 2.54 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
