
Actinomycetospora cinnamomea
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Actinomycetospora
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
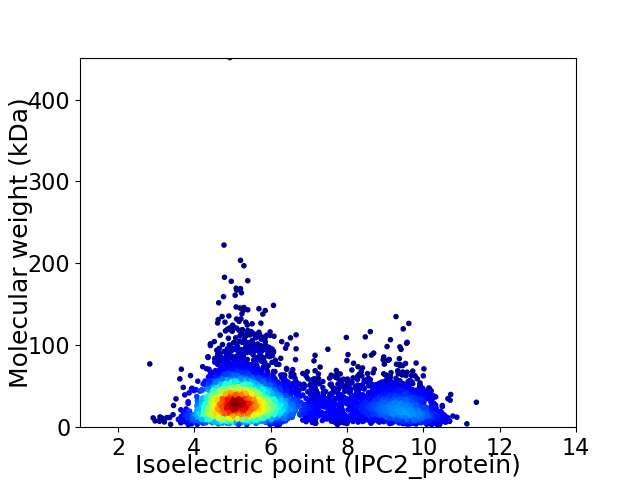
Virtual 2D-PAGE plot for 5694 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U1F6R2|A0A2U1F6R2_9PSEU Uncharacterized protein OS=Actinomycetospora cinnamomea OX=663609 GN=C8D89_11026 PE=4 SV=1
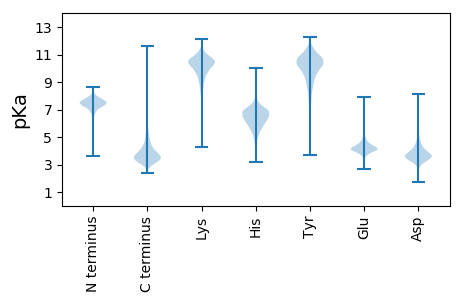
MM1 pKa = 7.83RR2 pKa = 11.84GRR4 pKa = 11.84RR5 pKa = 11.84SSRR8 pKa = 11.84SAAVVLAGCVGLAVMAPGLALAAPVPTSEE37 pKa = 4.94DD38 pKa = 3.19ACEE41 pKa = 3.89AAGGRR46 pKa = 11.84WTAGTCVPQRR56 pKa = 11.84TAPLTTSPAVEE67 pKa = 4.72DD68 pKa = 4.0EE69 pKa = 4.47TGGAATTPDD78 pKa = 3.91TPEE81 pKa = 3.68TAAPTTTTDD90 pKa = 3.53GLPSGGNPSSGTPATGGGSGGAGGAVGAGAGAQEE124 pKa = 4.68TPDD127 pKa = 3.62PTADD131 pKa = 3.58EE132 pKa = 4.83ASASADD138 pKa = 3.14QTTRR142 pKa = 11.84SPEE145 pKa = 4.09EE146 pKa = 3.86IGTQQSTARR155 pKa = 11.84ALIGPSALTSADD167 pKa = 3.36PAALVQDD174 pKa = 4.35LVEE177 pKa = 4.67GASGPGIDD185 pKa = 4.32FTNVPSPLTEE195 pKa = 4.55GGLPFEE201 pKa = 4.37NLEE204 pKa = 4.13IPEE207 pKa = 4.57LPADD211 pKa = 3.97GVFASPRR218 pKa = 11.84DD219 pKa = 3.47ACAYY223 pKa = 10.12LAGSVPAPEE232 pKa = 4.48QGAEE236 pKa = 3.91DD237 pKa = 3.46LAAAFEE243 pKa = 4.7SFCGALPEE251 pKa = 5.15DD252 pKa = 4.08FTEE255 pKa = 4.82FDD257 pKa = 3.33PTTIVGGLLDD267 pKa = 6.29LINKK271 pKa = 8.39LLKK274 pKa = 10.52ALPTPGNPIPSNPPAHH290 pKa = 6.39VPHH293 pKa = 7.25EE294 pKa = 4.23YY295 pKa = 9.5WGWWHH300 pKa = 6.29KK301 pKa = 10.96QYY303 pKa = 11.38DD304 pKa = 4.23VNCDD308 pKa = 3.14DD309 pKa = 3.55VTYY312 pKa = 11.01DD313 pKa = 3.4EE314 pKa = 5.55ANAILAWDD322 pKa = 3.71YY323 pKa = 11.78SDD325 pKa = 5.14PFNLDD330 pKa = 2.8GDD332 pKa = 4.36NDD334 pKa = 4.08GEE336 pKa = 4.1ACEE339 pKa = 5.02RR340 pKa = 11.84NWHH343 pKa = 6.39HH344 pKa = 7.68DD345 pKa = 3.17DD346 pKa = 3.6VEE348 pKa = 4.25YY349 pKa = 11.18VEE351 pKa = 4.21YY352 pKa = 10.92VEE354 pKa = 4.04YY355 pKa = 11.04VEE357 pKa = 4.19YY358 pKa = 11.08VEE360 pKa = 4.23YY361 pKa = 10.69DD362 pKa = 3.48SYY364 pKa = 11.7PVGGVATGDD373 pKa = 3.47GSTGADD379 pKa = 2.92SSAATGAEE387 pKa = 3.99VALAAGALSGLGLTGLVLVRR407 pKa = 11.84RR408 pKa = 11.84FARR411 pKa = 11.84QGG413 pKa = 3.07
MM1 pKa = 7.83RR2 pKa = 11.84GRR4 pKa = 11.84RR5 pKa = 11.84SSRR8 pKa = 11.84SAAVVLAGCVGLAVMAPGLALAAPVPTSEE37 pKa = 4.94DD38 pKa = 3.19ACEE41 pKa = 3.89AAGGRR46 pKa = 11.84WTAGTCVPQRR56 pKa = 11.84TAPLTTSPAVEE67 pKa = 4.72DD68 pKa = 4.0EE69 pKa = 4.47TGGAATTPDD78 pKa = 3.91TPEE81 pKa = 3.68TAAPTTTTDD90 pKa = 3.53GLPSGGNPSSGTPATGGGSGGAGGAVGAGAGAQEE124 pKa = 4.68TPDD127 pKa = 3.62PTADD131 pKa = 3.58EE132 pKa = 4.83ASASADD138 pKa = 3.14QTTRR142 pKa = 11.84SPEE145 pKa = 4.09EE146 pKa = 3.86IGTQQSTARR155 pKa = 11.84ALIGPSALTSADD167 pKa = 3.36PAALVQDD174 pKa = 4.35LVEE177 pKa = 4.67GASGPGIDD185 pKa = 4.32FTNVPSPLTEE195 pKa = 4.55GGLPFEE201 pKa = 4.37NLEE204 pKa = 4.13IPEE207 pKa = 4.57LPADD211 pKa = 3.97GVFASPRR218 pKa = 11.84DD219 pKa = 3.47ACAYY223 pKa = 10.12LAGSVPAPEE232 pKa = 4.48QGAEE236 pKa = 3.91DD237 pKa = 3.46LAAAFEE243 pKa = 4.7SFCGALPEE251 pKa = 5.15DD252 pKa = 4.08FTEE255 pKa = 4.82FDD257 pKa = 3.33PTTIVGGLLDD267 pKa = 6.29LINKK271 pKa = 8.39LLKK274 pKa = 10.52ALPTPGNPIPSNPPAHH290 pKa = 6.39VPHH293 pKa = 7.25EE294 pKa = 4.23YY295 pKa = 9.5WGWWHH300 pKa = 6.29KK301 pKa = 10.96QYY303 pKa = 11.38DD304 pKa = 4.23VNCDD308 pKa = 3.14DD309 pKa = 3.55VTYY312 pKa = 11.01DD313 pKa = 3.4EE314 pKa = 5.55ANAILAWDD322 pKa = 3.71YY323 pKa = 11.78SDD325 pKa = 5.14PFNLDD330 pKa = 2.8GDD332 pKa = 4.36NDD334 pKa = 4.08GEE336 pKa = 4.1ACEE339 pKa = 5.02RR340 pKa = 11.84NWHH343 pKa = 6.39HH344 pKa = 7.68DD345 pKa = 3.17DD346 pKa = 3.6VEE348 pKa = 4.25YY349 pKa = 11.18VEE351 pKa = 4.21YY352 pKa = 10.92VEE354 pKa = 4.04YY355 pKa = 11.04VEE357 pKa = 4.19YY358 pKa = 11.08VEE360 pKa = 4.23YY361 pKa = 10.69DD362 pKa = 3.48SYY364 pKa = 11.7PVGGVATGDD373 pKa = 3.47GSTGADD379 pKa = 2.92SSAATGAEE387 pKa = 3.99VALAAGALSGLGLTGLVLVRR407 pKa = 11.84RR408 pKa = 11.84FARR411 pKa = 11.84QGG413 pKa = 3.07
Molecular weight: 41.83 kDa
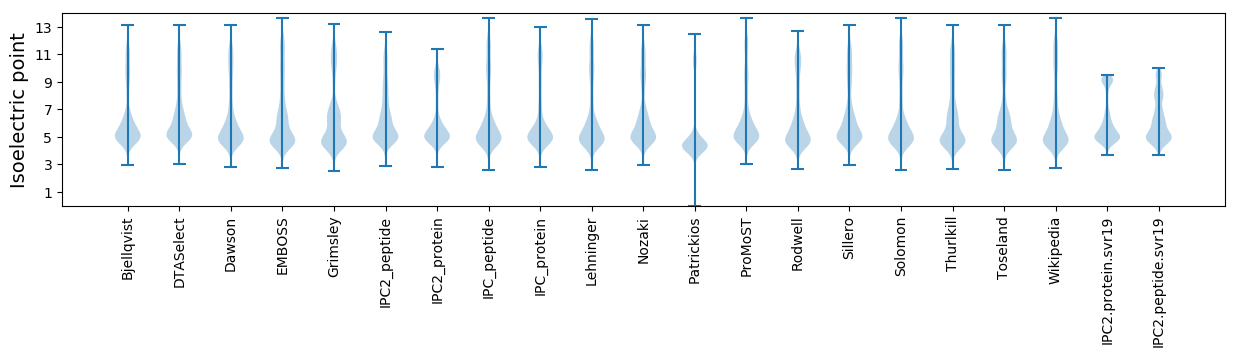
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U1FR97|A0A2U1FR97_9PSEU Methylmalonyl-CoA epimerase OS=Actinomycetospora cinnamomea OX=663609 GN=C8D89_101562 PE=3 SV=1
MM1 pKa = 7.74PATAGRR7 pKa = 11.84VPARR11 pKa = 11.84PRR13 pKa = 11.84VRR15 pKa = 11.84GPAAARR21 pKa = 11.84PSRR24 pKa = 11.84ARR26 pKa = 11.84PAARR30 pKa = 11.84PVAPPAPVRR39 pKa = 11.84PPARR43 pKa = 11.84VPGRR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84PTARR54 pKa = 11.84APAAPRR60 pKa = 11.84VPAPVPAPPRR70 pKa = 11.84ARR72 pKa = 11.84VPAPSPARR80 pKa = 11.84PPRR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84PRR89 pKa = 11.84PPRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84LPRR98 pKa = 11.84PVTPPAARR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84AHH110 pKa = 6.9RR111 pKa = 11.84APAGTVPVPVATARR125 pKa = 11.84VRR127 pKa = 11.84VARR130 pKa = 11.84RR131 pKa = 11.84GPVPSVPPSAVATSARR147 pKa = 11.84AEE149 pKa = 4.14VEE151 pKa = 4.01TAGTVVTARR160 pKa = 11.84PRR162 pKa = 11.84APVSSSSVRR171 pKa = 11.84PRR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84TQARR179 pKa = 11.84AVTARR184 pKa = 11.84RR185 pKa = 11.84SPRR188 pKa = 11.84ARR190 pKa = 11.84RR191 pKa = 11.84PRR193 pKa = 11.84SPGRR197 pKa = 11.84RR198 pKa = 11.84PRR200 pKa = 11.84ASATTPSGWAPAPRR214 pKa = 11.84RR215 pKa = 11.84ARR217 pKa = 11.84SVRR220 pKa = 11.84SRR222 pKa = 11.84VRR224 pKa = 11.84AARR227 pKa = 11.84VRR229 pKa = 11.84RR230 pKa = 11.84VTARR234 pKa = 11.84PAPTPRR240 pKa = 11.84RR241 pKa = 11.84ARR243 pKa = 11.84RR244 pKa = 11.84VTASVRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84TACPVPAAPVVPGRR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84ATCPRR276 pKa = 11.84VRR278 pKa = 11.84TPAA281 pKa = 3.42
MM1 pKa = 7.74PATAGRR7 pKa = 11.84VPARR11 pKa = 11.84PRR13 pKa = 11.84VRR15 pKa = 11.84GPAAARR21 pKa = 11.84PSRR24 pKa = 11.84ARR26 pKa = 11.84PAARR30 pKa = 11.84PVAPPAPVRR39 pKa = 11.84PPARR43 pKa = 11.84VPGRR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84PTARR54 pKa = 11.84APAAPRR60 pKa = 11.84VPAPVPAPPRR70 pKa = 11.84ARR72 pKa = 11.84VPAPSPARR80 pKa = 11.84PPRR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84RR87 pKa = 11.84PRR89 pKa = 11.84PPRR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84LPRR98 pKa = 11.84PVTPPAARR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84AHH110 pKa = 6.9RR111 pKa = 11.84APAGTVPVPVATARR125 pKa = 11.84VRR127 pKa = 11.84VARR130 pKa = 11.84RR131 pKa = 11.84GPVPSVPPSAVATSARR147 pKa = 11.84AEE149 pKa = 4.14VEE151 pKa = 4.01TAGTVVTARR160 pKa = 11.84PRR162 pKa = 11.84APVSSSSVRR171 pKa = 11.84PRR173 pKa = 11.84RR174 pKa = 11.84RR175 pKa = 11.84TQARR179 pKa = 11.84AVTARR184 pKa = 11.84RR185 pKa = 11.84SPRR188 pKa = 11.84ARR190 pKa = 11.84RR191 pKa = 11.84PRR193 pKa = 11.84SPGRR197 pKa = 11.84RR198 pKa = 11.84PRR200 pKa = 11.84ASATTPSGWAPAPRR214 pKa = 11.84RR215 pKa = 11.84ARR217 pKa = 11.84SVRR220 pKa = 11.84SRR222 pKa = 11.84VRR224 pKa = 11.84AARR227 pKa = 11.84VRR229 pKa = 11.84RR230 pKa = 11.84VTARR234 pKa = 11.84PAPTPRR240 pKa = 11.84RR241 pKa = 11.84ARR243 pKa = 11.84RR244 pKa = 11.84VTASVRR250 pKa = 11.84RR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84TACPVPAAPVVPGRR267 pKa = 11.84RR268 pKa = 11.84RR269 pKa = 11.84RR270 pKa = 11.84RR271 pKa = 11.84ATCPRR276 pKa = 11.84VRR278 pKa = 11.84TPAA281 pKa = 3.42
Molecular weight: 30.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1792867 |
27 |
4335 |
314.9 |
33.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.567 ± 0.058 | 0.728 ± 0.009 |
6.542 ± 0.027 | 5.646 ± 0.031 |
2.402 ± 0.017 | 9.841 ± 0.031 |
2.329 ± 0.017 | 2.65 ± 0.02 |
1.158 ± 0.019 | 10.272 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.559 ± 0.012 | 1.264 ± 0.014 |
6.626 ± 0.037 | 2.287 ± 0.018 |
8.886 ± 0.033 | 4.534 ± 0.021 |
5.878 ± 0.023 | 9.724 ± 0.04 |
1.471 ± 0.013 | 1.636 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
