
Tardiphaga sp. vice352
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Bradyrhizobiaceae; Tardiphaga; unclassified Tardiphaga
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
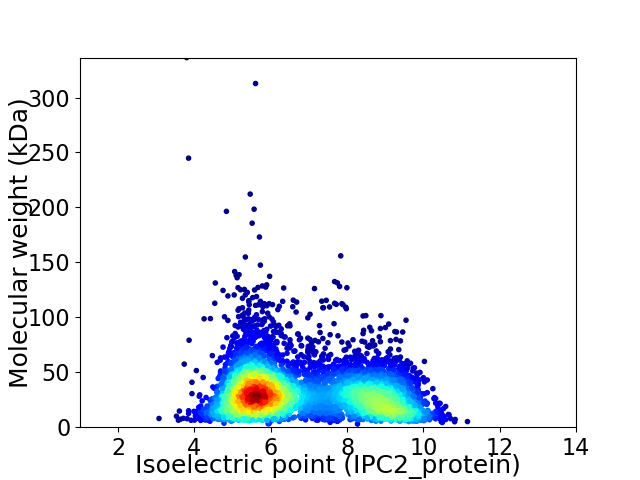
Virtual 2D-PAGE plot for 5026 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A515KMV7|A0A515KMV7_9BRAD Peptidase OS=Tardiphaga sp. vice352 OX=2592816 GN=FNL55_18430 PE=4 SV=1
MM1 pKa = 6.49TTADD5 pKa = 3.63TSQSKK10 pKa = 9.66YY11 pKa = 10.73AGPVWRR17 pKa = 11.84GVDD20 pKa = 4.13YY21 pKa = 11.33SPTWTAWVVGAGATQTGDD39 pKa = 3.05SDD41 pKa = 3.85FANDD45 pKa = 3.79AFQSLWAANFMAAPGGDD62 pKa = 3.14TSLPTDD68 pKa = 3.31NGNNYY73 pKa = 10.09RR74 pKa = 11.84NDD76 pKa = 3.51LQIIADD82 pKa = 3.68AGLNLVRR89 pKa = 11.84LYY91 pKa = 10.65NWDD94 pKa = 3.41MARR97 pKa = 11.84GTTATSNVGLDD108 pKa = 3.87HH109 pKa = 7.75INFLDD114 pKa = 3.67AAAGFGLKK122 pKa = 10.08VVVPVSDD129 pKa = 3.6WFLSDD134 pKa = 6.19DD135 pKa = 3.72PNAWNFTPAPLPSNYY150 pKa = 10.29DD151 pKa = 3.34FANAPAAIQTDD162 pKa = 4.2FSQFVASVTDD172 pKa = 3.56PTTGKK177 pKa = 9.14IHH179 pKa = 6.3SAVHH183 pKa = 6.32SIDD186 pKa = 3.32VGNEE190 pKa = 3.23GDD192 pKa = 4.14IGQGLNGQTTPSSFLYY208 pKa = 8.76RR209 pKa = 11.84TIWWIVNLHH218 pKa = 4.56QQINGGSPGPDD229 pKa = 2.76GSPAVNGATPVVPSTATFSNGDD251 pKa = 3.17QGLGIGSWFNCLIAGVAANQQTPTEE276 pKa = 4.21MDD278 pKa = 3.13GHH280 pKa = 6.98SSFDD284 pKa = 3.5VAVTGLKK291 pKa = 10.45AADD294 pKa = 4.04PAWEE298 pKa = 3.82QYY300 pKa = 10.64YY301 pKa = 10.93YY302 pKa = 11.34NSTNISQVSTATPFGNTLAATLALYY327 pKa = 10.56DD328 pKa = 4.29SGASPWPGADD338 pKa = 3.11CTVPLLLMEE347 pKa = 5.55LFTPNRR353 pKa = 11.84TAFPEE358 pKa = 4.29PADD361 pKa = 3.55QAVAAVGQVTDD372 pKa = 4.27LEE374 pKa = 4.83SYY376 pKa = 10.25LAQNSAGTASSSTYY390 pKa = 11.02LMGYY394 pKa = 10.38NYY396 pKa = 10.46FEE398 pKa = 5.28FNDD401 pKa = 3.73EE402 pKa = 4.18QQVKK406 pKa = 8.36LTGLYY411 pKa = 10.17QYY413 pKa = 11.26GATFNPAATGTTSVFYY429 pKa = 10.74SPYY432 pKa = 9.63QFPIMAFPVYY442 pKa = 10.57SLVATPGPNGSGTLIDD458 pKa = 5.54ALTQCVPGNIVAVFGEE474 pKa = 4.38TGNWQATFYY483 pKa = 9.68TGAPIPSWVTTGMSVWGANIPASTTVNVPTPGPGAVDD520 pKa = 3.23MTVVLVCAGSQASNPFTVEE539 pKa = 3.83TVTLSFYY546 pKa = 11.47
MM1 pKa = 6.49TTADD5 pKa = 3.63TSQSKK10 pKa = 9.66YY11 pKa = 10.73AGPVWRR17 pKa = 11.84GVDD20 pKa = 4.13YY21 pKa = 11.33SPTWTAWVVGAGATQTGDD39 pKa = 3.05SDD41 pKa = 3.85FANDD45 pKa = 3.79AFQSLWAANFMAAPGGDD62 pKa = 3.14TSLPTDD68 pKa = 3.31NGNNYY73 pKa = 10.09RR74 pKa = 11.84NDD76 pKa = 3.51LQIIADD82 pKa = 3.68AGLNLVRR89 pKa = 11.84LYY91 pKa = 10.65NWDD94 pKa = 3.41MARR97 pKa = 11.84GTTATSNVGLDD108 pKa = 3.87HH109 pKa = 7.75INFLDD114 pKa = 3.67AAAGFGLKK122 pKa = 10.08VVVPVSDD129 pKa = 3.6WFLSDD134 pKa = 6.19DD135 pKa = 3.72PNAWNFTPAPLPSNYY150 pKa = 10.29DD151 pKa = 3.34FANAPAAIQTDD162 pKa = 4.2FSQFVASVTDD172 pKa = 3.56PTTGKK177 pKa = 9.14IHH179 pKa = 6.3SAVHH183 pKa = 6.32SIDD186 pKa = 3.32VGNEE190 pKa = 3.23GDD192 pKa = 4.14IGQGLNGQTTPSSFLYY208 pKa = 8.76RR209 pKa = 11.84TIWWIVNLHH218 pKa = 4.56QQINGGSPGPDD229 pKa = 2.76GSPAVNGATPVVPSTATFSNGDD251 pKa = 3.17QGLGIGSWFNCLIAGVAANQQTPTEE276 pKa = 4.21MDD278 pKa = 3.13GHH280 pKa = 6.98SSFDD284 pKa = 3.5VAVTGLKK291 pKa = 10.45AADD294 pKa = 4.04PAWEE298 pKa = 3.82QYY300 pKa = 10.64YY301 pKa = 10.93YY302 pKa = 11.34NSTNISQVSTATPFGNTLAATLALYY327 pKa = 10.56DD328 pKa = 4.29SGASPWPGADD338 pKa = 3.11CTVPLLLMEE347 pKa = 5.55LFTPNRR353 pKa = 11.84TAFPEE358 pKa = 4.29PADD361 pKa = 3.55QAVAAVGQVTDD372 pKa = 4.27LEE374 pKa = 4.83SYY376 pKa = 10.25LAQNSAGTASSSTYY390 pKa = 11.02LMGYY394 pKa = 10.38NYY396 pKa = 10.46FEE398 pKa = 5.28FNDD401 pKa = 3.73EE402 pKa = 4.18QQVKK406 pKa = 8.36LTGLYY411 pKa = 10.17QYY413 pKa = 11.26GATFNPAATGTTSVFYY429 pKa = 10.74SPYY432 pKa = 9.63QFPIMAFPVYY442 pKa = 10.57SLVATPGPNGSGTLIDD458 pKa = 5.54ALTQCVPGNIVAVFGEE474 pKa = 4.38TGNWQATFYY483 pKa = 9.68TGAPIPSWVTTGMSVWGANIPASTTVNVPTPGPGAVDD520 pKa = 3.23MTVVLVCAGSQASNPFTVEE539 pKa = 3.83TVTLSFYY546 pKa = 11.47
Molecular weight: 57.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A515KQU1|A0A515KQU1_9BRAD 5'-methylthioadenosine nucleosidase OS=Tardiphaga sp. vice352 OX=2592816 GN=FNL55_24465 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATVGGRR28 pKa = 11.84KK29 pKa = 9.08VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATVGGRR28 pKa = 11.84KK29 pKa = 9.08VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1561869 |
26 |
3211 |
310.8 |
33.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.515 ± 0.045 | 0.827 ± 0.009 |
5.67 ± 0.027 | 5.155 ± 0.03 |
3.751 ± 0.022 | 8.321 ± 0.033 |
1.996 ± 0.017 | 5.495 ± 0.023 |
3.737 ± 0.035 | 9.856 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.606 ± 0.018 | 2.754 ± 0.019 |
5.194 ± 0.031 | 3.207 ± 0.017 |
6.766 ± 0.036 | 5.645 ± 0.024 |
5.479 ± 0.024 | 7.468 ± 0.025 |
1.311 ± 0.015 | 2.247 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
