
Pseudohaliea rubra DSM 19751
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Halieaceae; Pseudohaliea; Pseudohaliea rubra
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
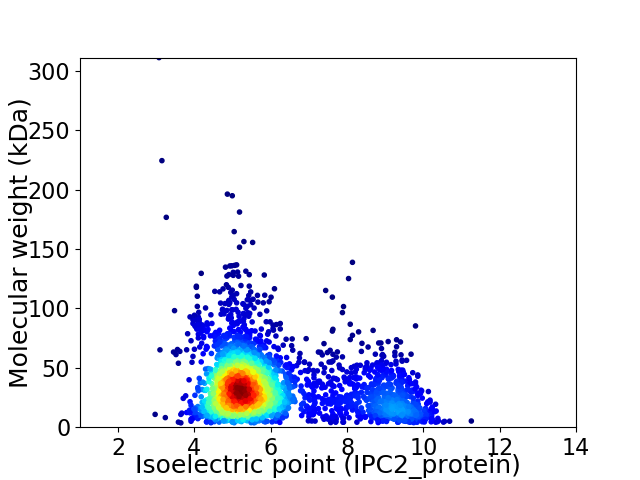
Virtual 2D-PAGE plot for 2860 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
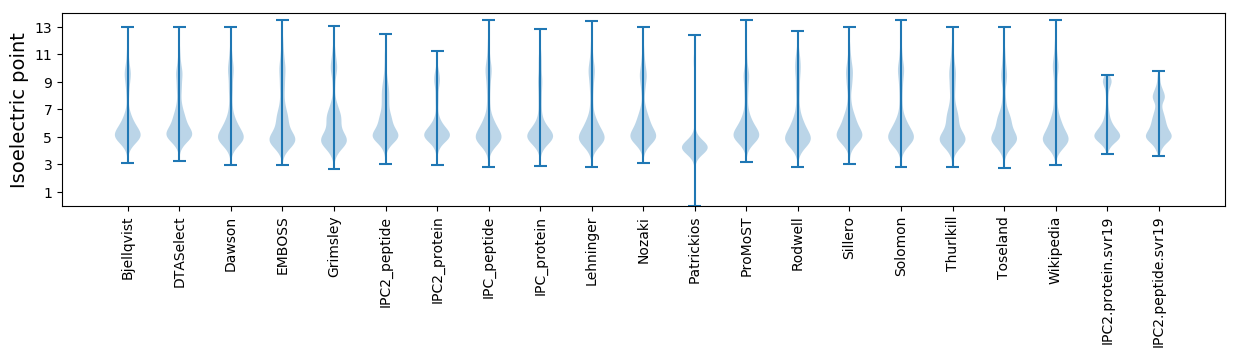
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A095VN58|A0A095VN58_9GAMM Dipeptide transport ATP-binding protein DppD OS=Pseudohaliea rubra DSM 19751 OX=1265313 GN=HRUBRA_02556 PE=4 SV=1
MM1 pKa = 6.75SHH3 pKa = 5.89WVATLGRR10 pKa = 11.84RR11 pKa = 11.84LAALGSMLVLLTACGGGGGGGGFLSDD37 pKa = 5.35DD38 pKa = 3.64GTGNNPLTITTGTLPDD54 pKa = 3.73VASTPYY60 pKa = 9.64STVLEE65 pKa = 4.47ASGGTPPYY73 pKa = 10.1SWEE76 pKa = 4.7LIDD79 pKa = 5.98DD80 pKa = 4.56GGTGFTINSEE90 pKa = 4.37GIFRR94 pKa = 11.84GDD96 pKa = 3.61ALPPAGTYY104 pKa = 10.46GVTISVSDD112 pKa = 3.6ANGRR116 pKa = 11.84DD117 pKa = 3.41TRR119 pKa = 11.84QSFTLTVTTEE129 pKa = 3.87NALSIATTALPQAIEE144 pKa = 4.11GLDD147 pKa = 3.48YY148 pKa = 10.58TALLDD153 pKa = 3.87ASGGAEE159 pKa = 4.02PYY161 pKa = 9.73RR162 pKa = 11.84WSVVNDD168 pKa = 3.48GGTGFGINADD178 pKa = 3.63GVLTGTAPDD187 pKa = 3.56SGDD190 pKa = 3.3YY191 pKa = 11.17GITLEE196 pKa = 4.44VTDD199 pKa = 4.61EE200 pKa = 4.09ADD202 pKa = 3.36SRR204 pKa = 11.84DD205 pKa = 3.76RR206 pKa = 11.84QSFILTVTGDD216 pKa = 3.6TPQPLTIATEE226 pKa = 4.12TLPTAEE232 pKa = 4.46EE233 pKa = 4.47GQTYY237 pKa = 10.47SAILQAAGGQGDD249 pKa = 4.42YY250 pKa = 11.28LWTLVDD256 pKa = 4.11AGGTGLDD263 pKa = 3.8LRR265 pKa = 11.84DD266 pKa = 4.47DD267 pKa = 4.3GVLSGTVPAEE277 pKa = 3.65GSYY280 pKa = 10.98GITVAVADD288 pKa = 4.03NVRR291 pKa = 11.84EE292 pKa = 4.06VSKK295 pKa = 11.21SLVLTVSAEE304 pKa = 4.01GDD306 pKa = 3.78PLTLGTTSLPGATVGEE322 pKa = 4.62RR323 pKa = 11.84YY324 pKa = 9.64AAVLEE329 pKa = 4.29ATGGSRR335 pKa = 11.84DD336 pKa = 3.8YY337 pKa = 11.26SWQLVSSGDD346 pKa = 3.57SGLSLSSAGVLSGTPSAVGTYY367 pKa = 10.18GIVFRR372 pKa = 11.84VSDD375 pKa = 3.99GQDD378 pKa = 3.0TVQGALTLVVDD389 pKa = 4.01TFEE392 pKa = 4.64PFVPLAVVTEE402 pKa = 4.14TLPAANGQIYY412 pKa = 10.15AATLEE417 pKa = 4.58AEE419 pKa = 4.87GGTPPYY425 pKa = 10.9SWTLVNDD432 pKa = 4.33GGSGLSLSTSGSLSGTSPAVGTYY455 pKa = 10.44GLTVSVADD463 pKa = 3.81SAGGSDD469 pKa = 3.42TAALTLEE476 pKa = 4.34VDD478 pKa = 3.85GSDD481 pKa = 3.05ATTLEE486 pKa = 4.73IISTDD491 pKa = 3.72LGTATQGEE499 pKa = 5.15TYY501 pKa = 10.9SFILRR506 pKa = 11.84ASGGPTGDD514 pKa = 3.77YY515 pKa = 8.37TWSKK519 pKa = 10.73VEE521 pKa = 4.23VVPSLPTIEE530 pKa = 4.63IGSDD534 pKa = 3.12SFEE537 pKa = 4.34DD538 pKa = 4.13ARR540 pKa = 11.84LTLDD544 pKa = 3.98PDD546 pKa = 3.85TGVLTWFSGDD556 pKa = 3.13IEE558 pKa = 4.7NGVLTGEE565 pKa = 4.39CTTDD569 pKa = 2.94YY570 pKa = 10.9SYY572 pKa = 10.82TVQVTDD578 pKa = 4.36AGPPASSDD586 pKa = 3.36VRR588 pKa = 11.84TLPLTAALDD597 pKa = 4.29PNDD600 pKa = 5.12PDD602 pKa = 3.94CQQ604 pKa = 3.59
MM1 pKa = 6.75SHH3 pKa = 5.89WVATLGRR10 pKa = 11.84RR11 pKa = 11.84LAALGSMLVLLTACGGGGGGGGFLSDD37 pKa = 5.35DD38 pKa = 3.64GTGNNPLTITTGTLPDD54 pKa = 3.73VASTPYY60 pKa = 9.64STVLEE65 pKa = 4.47ASGGTPPYY73 pKa = 10.1SWEE76 pKa = 4.7LIDD79 pKa = 5.98DD80 pKa = 4.56GGTGFTINSEE90 pKa = 4.37GIFRR94 pKa = 11.84GDD96 pKa = 3.61ALPPAGTYY104 pKa = 10.46GVTISVSDD112 pKa = 3.6ANGRR116 pKa = 11.84DD117 pKa = 3.41TRR119 pKa = 11.84QSFTLTVTTEE129 pKa = 3.87NALSIATTALPQAIEE144 pKa = 4.11GLDD147 pKa = 3.48YY148 pKa = 10.58TALLDD153 pKa = 3.87ASGGAEE159 pKa = 4.02PYY161 pKa = 9.73RR162 pKa = 11.84WSVVNDD168 pKa = 3.48GGTGFGINADD178 pKa = 3.63GVLTGTAPDD187 pKa = 3.56SGDD190 pKa = 3.3YY191 pKa = 11.17GITLEE196 pKa = 4.44VTDD199 pKa = 4.61EE200 pKa = 4.09ADD202 pKa = 3.36SRR204 pKa = 11.84DD205 pKa = 3.76RR206 pKa = 11.84QSFILTVTGDD216 pKa = 3.6TPQPLTIATEE226 pKa = 4.12TLPTAEE232 pKa = 4.46EE233 pKa = 4.47GQTYY237 pKa = 10.47SAILQAAGGQGDD249 pKa = 4.42YY250 pKa = 11.28LWTLVDD256 pKa = 4.11AGGTGLDD263 pKa = 3.8LRR265 pKa = 11.84DD266 pKa = 4.47DD267 pKa = 4.3GVLSGTVPAEE277 pKa = 3.65GSYY280 pKa = 10.98GITVAVADD288 pKa = 4.03NVRR291 pKa = 11.84EE292 pKa = 4.06VSKK295 pKa = 11.21SLVLTVSAEE304 pKa = 4.01GDD306 pKa = 3.78PLTLGTTSLPGATVGEE322 pKa = 4.62RR323 pKa = 11.84YY324 pKa = 9.64AAVLEE329 pKa = 4.29ATGGSRR335 pKa = 11.84DD336 pKa = 3.8YY337 pKa = 11.26SWQLVSSGDD346 pKa = 3.57SGLSLSSAGVLSGTPSAVGTYY367 pKa = 10.18GIVFRR372 pKa = 11.84VSDD375 pKa = 3.99GQDD378 pKa = 3.0TVQGALTLVVDD389 pKa = 4.01TFEE392 pKa = 4.64PFVPLAVVTEE402 pKa = 4.14TLPAANGQIYY412 pKa = 10.15AATLEE417 pKa = 4.58AEE419 pKa = 4.87GGTPPYY425 pKa = 10.9SWTLVNDD432 pKa = 4.33GGSGLSLSTSGSLSGTSPAVGTYY455 pKa = 10.44GLTVSVADD463 pKa = 3.81SAGGSDD469 pKa = 3.42TAALTLEE476 pKa = 4.34VDD478 pKa = 3.85GSDD481 pKa = 3.05ATTLEE486 pKa = 4.73IISTDD491 pKa = 3.72LGTATQGEE499 pKa = 5.15TYY501 pKa = 10.9SFILRR506 pKa = 11.84ASGGPTGDD514 pKa = 3.77YY515 pKa = 8.37TWSKK519 pKa = 10.73VEE521 pKa = 4.23VVPSLPTIEE530 pKa = 4.63IGSDD534 pKa = 3.12SFEE537 pKa = 4.34DD538 pKa = 4.13ARR540 pKa = 11.84LTLDD544 pKa = 3.98PDD546 pKa = 3.85TGVLTWFSGDD556 pKa = 3.13IEE558 pKa = 4.7NGVLTGEE565 pKa = 4.39CTTDD569 pKa = 2.94YY570 pKa = 10.9SYY572 pKa = 10.82TVQVTDD578 pKa = 4.36AGPPASSDD586 pKa = 3.36VRR588 pKa = 11.84TLPLTAALDD597 pKa = 4.29PNDD600 pKa = 5.12PDD602 pKa = 3.94CQQ604 pKa = 3.59
Molecular weight: 61.14 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A095VSJ5|A0A095VSJ5_9GAMM Permease of the drug/metabolite transporter (DMT) superfamily OS=Pseudohaliea rubra DSM 19751 OX=1265313 GN=HRUBRA_01389 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.35NGRR28 pKa = 11.84LVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.35NGRR28 pKa = 11.84LVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
961170 |
37 |
3013 |
336.1 |
36.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.862 ± 0.062 | 0.978 ± 0.016 |
6.007 ± 0.048 | 6.336 ± 0.039 |
3.554 ± 0.03 | 8.794 ± 0.043 |
2.078 ± 0.024 | 4.102 ± 0.032 |
2.189 ± 0.028 | 11.471 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.024 ± 0.02 | 2.43 ± 0.026 |
5.155 ± 0.027 | 3.146 ± 0.026 |
7.679 ± 0.054 | 5.037 ± 0.032 |
4.942 ± 0.045 | 7.333 ± 0.038 |
1.356 ± 0.019 | 2.528 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
