
Petrochirus diogenes giant hermit crab associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.58
Get precalculated fractions of proteins
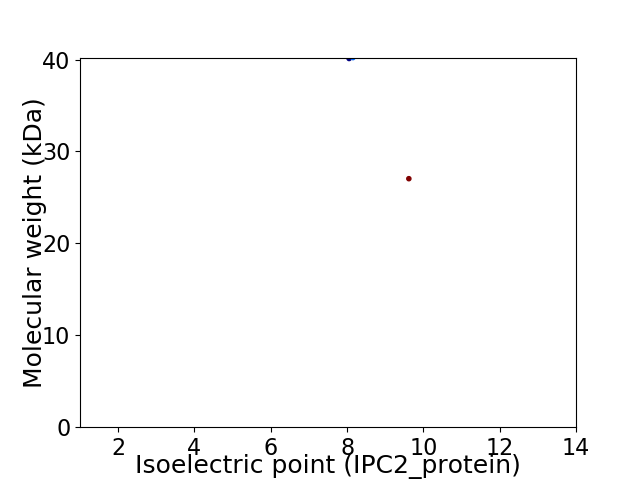
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RLL5|A0A0K1RLL5_9CIRC Putative replication initiation protein OS=Petrochirus diogenes giant hermit crab associated circular virus OX=1692261 PE=4 SV=1
MM1 pKa = 6.99EE2 pKa = 5.46GKK4 pKa = 9.64RR5 pKa = 11.84VRR7 pKa = 11.84YY8 pKa = 9.22RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.17CITINNEE18 pKa = 3.7SDD20 pKa = 2.96NKK22 pKa = 10.59RR23 pKa = 11.84ILGLFLEE30 pKa = 5.08DD31 pKa = 4.48GIKK34 pKa = 10.34KK35 pKa = 10.25GVIGVEE41 pKa = 4.11TAPTTGTKK49 pKa = 9.96HH50 pKa = 4.62LQGYY54 pKa = 9.09IEE56 pKa = 5.03YY57 pKa = 7.41PTQRR61 pKa = 11.84DD62 pKa = 2.95WSSIKK67 pKa = 10.17KK68 pKa = 9.0VCPNAHH74 pKa = 6.13IEE76 pKa = 4.11AAKK79 pKa = 8.95GTSRR83 pKa = 11.84QNYY86 pKa = 7.03EE87 pKa = 3.86YY88 pKa = 9.51CTKK91 pKa = 10.18EE92 pKa = 3.83GNFVVHH98 pKa = 6.21GHH100 pKa = 6.2FEE102 pKa = 3.81TGKK105 pKa = 10.04RR106 pKa = 11.84KK107 pKa = 9.87RR108 pKa = 11.84RR109 pKa = 11.84DD110 pKa = 3.47YY111 pKa = 11.46SLSEE115 pKa = 3.92YY116 pKa = 10.21VKK118 pKa = 10.61QVLQDD123 pKa = 3.71EE124 pKa = 5.02LPNDD128 pKa = 3.42STFIRR133 pKa = 11.84NYY135 pKa = 10.61EE136 pKa = 4.27KK137 pKa = 10.55IQTHH141 pKa = 6.17AKK143 pKa = 9.62LVSSRR148 pKa = 11.84QEE150 pKa = 3.65KK151 pKa = 10.03VRR153 pKa = 11.84LFGEE157 pKa = 4.54LSCCKK162 pKa = 8.76VTSWQRR168 pKa = 11.84KK169 pKa = 8.0AIEE172 pKa = 4.13SLFLQGKK179 pKa = 9.51RR180 pKa = 11.84EE181 pKa = 3.96VLWIYY186 pKa = 10.62EE187 pKa = 4.34SVGGKK192 pKa = 10.19GKK194 pKa = 8.24TFLATILEE202 pKa = 4.4VVYY205 pKa = 10.56GFTRR209 pKa = 11.84FDD211 pKa = 4.77GITKK215 pKa = 10.36SRR217 pKa = 11.84DD218 pKa = 2.98IALLIPPIPKK228 pKa = 10.15GFVFDD233 pKa = 3.61VTRR236 pKa = 11.84DD237 pKa = 3.52DD238 pKa = 4.83ASNFSYY244 pKa = 9.44NTLEE248 pKa = 4.04QVKK251 pKa = 9.94NGYY254 pKa = 9.36VMSGKK259 pKa = 10.55YY260 pKa = 10.08GGSQHH265 pKa = 7.06LFRR268 pKa = 11.84PVPVIVLSNFEE279 pKa = 4.93PIRR282 pKa = 11.84SSLSEE287 pKa = 4.27DD288 pKa = 2.76RR289 pKa = 11.84WKK291 pKa = 10.79VWNIEE296 pKa = 3.93NASNEE301 pKa = 4.15KK302 pKa = 10.68KK303 pKa = 10.33EE304 pKa = 4.11DD305 pKa = 3.57LPEE308 pKa = 4.21EE309 pKa = 4.37GKK311 pKa = 10.25IPPKK315 pKa = 10.4AVLFEE320 pKa = 4.16EE321 pKa = 4.81EE322 pKa = 4.41EE323 pKa = 4.24NKK325 pKa = 10.37EE326 pKa = 4.03NQPNAEE332 pKa = 4.4ASNSQQSNKK341 pKa = 10.13DD342 pKa = 3.58VEE344 pKa = 4.31QHH346 pKa = 6.07RR347 pKa = 11.84EE348 pKa = 3.66KK349 pKa = 11.16
MM1 pKa = 6.99EE2 pKa = 5.46GKK4 pKa = 9.64RR5 pKa = 11.84VRR7 pKa = 11.84YY8 pKa = 9.22RR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.17CITINNEE18 pKa = 3.7SDD20 pKa = 2.96NKK22 pKa = 10.59RR23 pKa = 11.84ILGLFLEE30 pKa = 5.08DD31 pKa = 4.48GIKK34 pKa = 10.34KK35 pKa = 10.25GVIGVEE41 pKa = 4.11TAPTTGTKK49 pKa = 9.96HH50 pKa = 4.62LQGYY54 pKa = 9.09IEE56 pKa = 5.03YY57 pKa = 7.41PTQRR61 pKa = 11.84DD62 pKa = 2.95WSSIKK67 pKa = 10.17KK68 pKa = 9.0VCPNAHH74 pKa = 6.13IEE76 pKa = 4.11AAKK79 pKa = 8.95GTSRR83 pKa = 11.84QNYY86 pKa = 7.03EE87 pKa = 3.86YY88 pKa = 9.51CTKK91 pKa = 10.18EE92 pKa = 3.83GNFVVHH98 pKa = 6.21GHH100 pKa = 6.2FEE102 pKa = 3.81TGKK105 pKa = 10.04RR106 pKa = 11.84KK107 pKa = 9.87RR108 pKa = 11.84RR109 pKa = 11.84DD110 pKa = 3.47YY111 pKa = 11.46SLSEE115 pKa = 3.92YY116 pKa = 10.21VKK118 pKa = 10.61QVLQDD123 pKa = 3.71EE124 pKa = 5.02LPNDD128 pKa = 3.42STFIRR133 pKa = 11.84NYY135 pKa = 10.61EE136 pKa = 4.27KK137 pKa = 10.55IQTHH141 pKa = 6.17AKK143 pKa = 9.62LVSSRR148 pKa = 11.84QEE150 pKa = 3.65KK151 pKa = 10.03VRR153 pKa = 11.84LFGEE157 pKa = 4.54LSCCKK162 pKa = 8.76VTSWQRR168 pKa = 11.84KK169 pKa = 8.0AIEE172 pKa = 4.13SLFLQGKK179 pKa = 9.51RR180 pKa = 11.84EE181 pKa = 3.96VLWIYY186 pKa = 10.62EE187 pKa = 4.34SVGGKK192 pKa = 10.19GKK194 pKa = 8.24TFLATILEE202 pKa = 4.4VVYY205 pKa = 10.56GFTRR209 pKa = 11.84FDD211 pKa = 4.77GITKK215 pKa = 10.36SRR217 pKa = 11.84DD218 pKa = 2.98IALLIPPIPKK228 pKa = 10.15GFVFDD233 pKa = 3.61VTRR236 pKa = 11.84DD237 pKa = 3.52DD238 pKa = 4.83ASNFSYY244 pKa = 9.44NTLEE248 pKa = 4.04QVKK251 pKa = 9.94NGYY254 pKa = 9.36VMSGKK259 pKa = 10.55YY260 pKa = 10.08GGSQHH265 pKa = 7.06LFRR268 pKa = 11.84PVPVIVLSNFEE279 pKa = 4.93PIRR282 pKa = 11.84SSLSEE287 pKa = 4.27DD288 pKa = 2.76RR289 pKa = 11.84WKK291 pKa = 10.79VWNIEE296 pKa = 3.93NASNEE301 pKa = 4.15KK302 pKa = 10.68KK303 pKa = 10.33EE304 pKa = 4.11DD305 pKa = 3.57LPEE308 pKa = 4.21EE309 pKa = 4.37GKK311 pKa = 10.25IPPKK315 pKa = 10.4AVLFEE320 pKa = 4.16EE321 pKa = 4.81EE322 pKa = 4.41EE323 pKa = 4.24NKK325 pKa = 10.37EE326 pKa = 4.03NQPNAEE332 pKa = 4.4ASNSQQSNKK341 pKa = 10.13DD342 pKa = 3.58VEE344 pKa = 4.31QHH346 pKa = 6.07RR347 pKa = 11.84EE348 pKa = 3.66KK349 pKa = 11.16
Molecular weight: 40.12 kDa
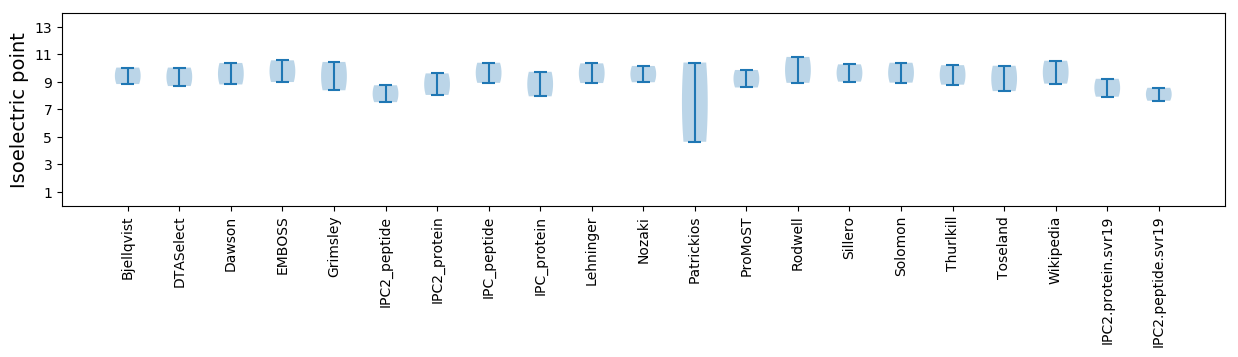
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RLL5|A0A0K1RLL5_9CIRC Putative replication initiation protein OS=Petrochirus diogenes giant hermit crab associated circular virus OX=1692261 PE=4 SV=1
MM1 pKa = 7.42PRR3 pKa = 11.84MRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 8.73KK8 pKa = 8.33TFRR11 pKa = 11.84KK12 pKa = 9.26KK13 pKa = 10.75GKK15 pKa = 6.67FHH17 pKa = 7.05RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 8.13YY22 pKa = 9.25SKK24 pKa = 10.38KK25 pKa = 9.59RR26 pKa = 11.84RR27 pKa = 11.84TRR29 pKa = 11.84RR30 pKa = 11.84INLMQKK36 pKa = 9.12QVIHH40 pKa = 6.38NKK42 pKa = 6.64VTRR45 pKa = 11.84TLNNIVKK52 pKa = 9.97SEE54 pKa = 3.98WNTPTEE60 pKa = 3.99LRR62 pKa = 11.84LSIMDD67 pKa = 5.15LLTTTEE73 pKa = 3.95QEE75 pKa = 3.88VFKK78 pKa = 10.97AFKK81 pKa = 9.29YY82 pKa = 9.59YY83 pKa = 10.56KK84 pKa = 9.38IRR86 pKa = 11.84GVKK89 pKa = 9.97VEE91 pKa = 5.21FKK93 pKa = 9.78CTPPVAAIAPQAVLTSDD110 pKa = 2.84IVHH113 pKa = 6.84IPNVLPIMYY122 pKa = 9.59CWEE125 pKa = 4.1RR126 pKa = 11.84NNDD129 pKa = 2.76VDD131 pKa = 4.03YY132 pKa = 10.28TISRR136 pKa = 11.84MKK138 pKa = 10.37EE139 pKa = 3.61NPNRR143 pKa = 11.84KK144 pKa = 8.87QFDD147 pKa = 3.87PYY149 pKa = 10.96KK150 pKa = 9.91GYY152 pKa = 10.81CKK154 pKa = 9.85TYY156 pKa = 10.43RR157 pKa = 11.84GLLPEE162 pKa = 4.62FQDD165 pKa = 3.84KK166 pKa = 11.07SGNNLFLNKK175 pKa = 9.97KK176 pKa = 8.74SWISSADD183 pKa = 3.16TTYY186 pKa = 11.21LYY188 pKa = 11.27GRR190 pKa = 11.84FIFMPNIISTDD201 pKa = 3.44ALIVNYY207 pKa = 10.42NFDD210 pKa = 3.77IDD212 pKa = 3.78LTIYY216 pKa = 10.84YY217 pKa = 9.81SLKK220 pKa = 8.54TLQYY224 pKa = 10.05TII226 pKa = 4.6
MM1 pKa = 7.42PRR3 pKa = 11.84MRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 8.73KK8 pKa = 8.33TFRR11 pKa = 11.84KK12 pKa = 9.26KK13 pKa = 10.75GKK15 pKa = 6.67FHH17 pKa = 7.05RR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84YY21 pKa = 8.13YY22 pKa = 9.25SKK24 pKa = 10.38KK25 pKa = 9.59RR26 pKa = 11.84RR27 pKa = 11.84TRR29 pKa = 11.84RR30 pKa = 11.84INLMQKK36 pKa = 9.12QVIHH40 pKa = 6.38NKK42 pKa = 6.64VTRR45 pKa = 11.84TLNNIVKK52 pKa = 9.97SEE54 pKa = 3.98WNTPTEE60 pKa = 3.99LRR62 pKa = 11.84LSIMDD67 pKa = 5.15LLTTTEE73 pKa = 3.95QEE75 pKa = 3.88VFKK78 pKa = 10.97AFKK81 pKa = 9.29YY82 pKa = 9.59YY83 pKa = 10.56KK84 pKa = 9.38IRR86 pKa = 11.84GVKK89 pKa = 9.97VEE91 pKa = 5.21FKK93 pKa = 9.78CTPPVAAIAPQAVLTSDD110 pKa = 2.84IVHH113 pKa = 6.84IPNVLPIMYY122 pKa = 9.59CWEE125 pKa = 4.1RR126 pKa = 11.84NNDD129 pKa = 2.76VDD131 pKa = 4.03YY132 pKa = 10.28TISRR136 pKa = 11.84MKK138 pKa = 10.37EE139 pKa = 3.61NPNRR143 pKa = 11.84KK144 pKa = 8.87QFDD147 pKa = 3.87PYY149 pKa = 10.96KK150 pKa = 9.91GYY152 pKa = 10.81CKK154 pKa = 9.85TYY156 pKa = 10.43RR157 pKa = 11.84GLLPEE162 pKa = 4.62FQDD165 pKa = 3.84KK166 pKa = 11.07SGNNLFLNKK175 pKa = 9.97KK176 pKa = 8.74SWISSADD183 pKa = 3.16TTYY186 pKa = 11.21LYY188 pKa = 11.27GRR190 pKa = 11.84FIFMPNIISTDD201 pKa = 3.44ALIVNYY207 pKa = 10.42NFDD210 pKa = 3.77IDD212 pKa = 3.78LTIYY216 pKa = 10.84YY217 pKa = 9.81SLKK220 pKa = 8.54TLQYY224 pKa = 10.05TII226 pKa = 4.6
Molecular weight: 27.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
575 |
226 |
349 |
287.5 |
33.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.478 ± 0.211 | 1.391 ± 0.035 |
4.174 ± 0.139 | 7.304 ± 2.081 |
4.522 ± 0.191 | 5.217 ± 1.417 |
1.739 ± 0.228 | 6.783 ± 0.653 |
9.565 ± 0.338 | 6.957 ± 0.313 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.565 ± 0.847 | 6.261 ± 0.453 |
4.348 ± 0.287 | 3.826 ± 0.403 |
7.13 ± 0.706 | 6.435 ± 0.867 |
6.435 ± 1.09 | 6.435 ± 0.622 |
1.391 ± 0.035 | 5.043 ± 0.881 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
