
Rhodobacter sp. 24-YEA-8
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Rhodobacter; unclassified Rhodobacter
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
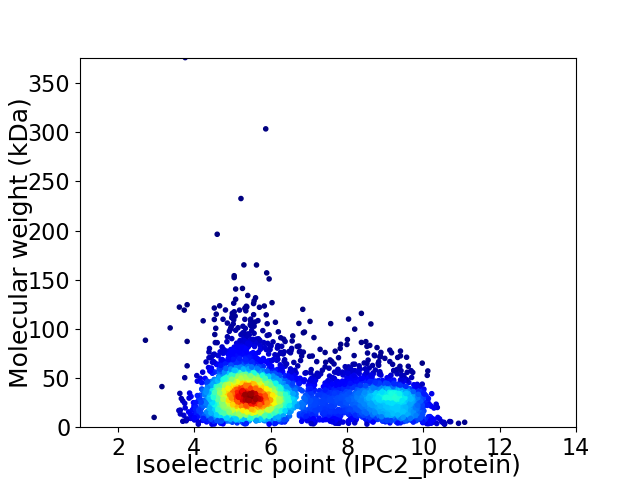
Virtual 2D-PAGE plot for 4521 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H4TB81|A0A1H4TB81_9RHOB ElaA protein OS=Rhodobacter sp. 24-YEA-8 OX=1884310 GN=SAMN05519105_2766 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 10.18KK3 pKa = 10.52LLIATTALVAGASAAAADD21 pKa = 4.1VTISGYY27 pKa = 11.03GRR29 pKa = 11.84FGLQYY34 pKa = 10.16IEE36 pKa = 4.89DD37 pKa = 3.83RR38 pKa = 11.84GVTPGSGIAGTEE50 pKa = 4.06EE51 pKa = 3.86EE52 pKa = 4.33TQIAGRR58 pKa = 11.84LRR60 pKa = 11.84LNIDD64 pKa = 3.16ATTEE68 pKa = 3.83TDD70 pKa = 3.26VGVTFGARR78 pKa = 11.84LRR80 pKa = 11.84LQNDD84 pKa = 4.28LGNDD88 pKa = 3.39TTVGNKK94 pKa = 9.28ARR96 pKa = 11.84FSVGYY101 pKa = 9.97EE102 pKa = 3.78GLTVQVGNVDD112 pKa = 3.92TAWDD116 pKa = 4.15SVRR119 pKa = 11.84LTYY122 pKa = 10.55DD123 pKa = 2.91SEE125 pKa = 4.34MGFDD129 pKa = 4.12DD130 pKa = 5.46SSFGDD135 pKa = 3.52AQNGFFAYY143 pKa = 9.86NSKK146 pKa = 10.83GADD149 pKa = 3.22SNQNSNYY156 pKa = 10.22AGVAAFYY163 pKa = 10.84SIADD167 pKa = 3.44VNLYY171 pKa = 10.16FSYY174 pKa = 11.23VDD176 pKa = 3.64PDD178 pKa = 3.42QTAKK182 pKa = 10.75DD183 pKa = 3.77LSIVPGLLANDD194 pKa = 3.92EE195 pKa = 4.73EE196 pKa = 5.12IGLAADD202 pKa = 4.02WSNGQISVAAGINLSSQGIKK222 pKa = 10.98DD223 pKa = 3.43NDD225 pKa = 3.92DD226 pKa = 3.72YY227 pKa = 11.7FIGAAYY233 pKa = 10.66NFGDD237 pKa = 3.85GNVGLNYY244 pKa = 10.11YY245 pKa = 10.25DD246 pKa = 4.31YY247 pKa = 9.38NTAAGGVSDD256 pKa = 3.9STQVTLYY263 pKa = 11.25GNYY266 pKa = 9.26TFGATTLKK274 pKa = 10.68AYY276 pKa = 10.63VSDD279 pKa = 3.83WDD281 pKa = 3.68RR282 pKa = 11.84AGYY285 pKa = 7.86DD286 pKa = 3.12TAYY289 pKa = 10.66GIGADD294 pKa = 3.68YY295 pKa = 11.25SLGEE299 pKa = 4.17GARR302 pKa = 11.84ISGSIQSGFGDD313 pKa = 3.4AAGANNGTKK322 pKa = 10.19ADD324 pKa = 3.75LGVRR328 pKa = 11.84FDD330 pKa = 3.67FF331 pKa = 5.43
MM1 pKa = 7.61KK2 pKa = 10.18KK3 pKa = 10.52LLIATTALVAGASAAAADD21 pKa = 4.1VTISGYY27 pKa = 11.03GRR29 pKa = 11.84FGLQYY34 pKa = 10.16IEE36 pKa = 4.89DD37 pKa = 3.83RR38 pKa = 11.84GVTPGSGIAGTEE50 pKa = 4.06EE51 pKa = 3.86EE52 pKa = 4.33TQIAGRR58 pKa = 11.84LRR60 pKa = 11.84LNIDD64 pKa = 3.16ATTEE68 pKa = 3.83TDD70 pKa = 3.26VGVTFGARR78 pKa = 11.84LRR80 pKa = 11.84LQNDD84 pKa = 4.28LGNDD88 pKa = 3.39TTVGNKK94 pKa = 9.28ARR96 pKa = 11.84FSVGYY101 pKa = 9.97EE102 pKa = 3.78GLTVQVGNVDD112 pKa = 3.92TAWDD116 pKa = 4.15SVRR119 pKa = 11.84LTYY122 pKa = 10.55DD123 pKa = 2.91SEE125 pKa = 4.34MGFDD129 pKa = 4.12DD130 pKa = 5.46SSFGDD135 pKa = 3.52AQNGFFAYY143 pKa = 9.86NSKK146 pKa = 10.83GADD149 pKa = 3.22SNQNSNYY156 pKa = 10.22AGVAAFYY163 pKa = 10.84SIADD167 pKa = 3.44VNLYY171 pKa = 10.16FSYY174 pKa = 11.23VDD176 pKa = 3.64PDD178 pKa = 3.42QTAKK182 pKa = 10.75DD183 pKa = 3.77LSIVPGLLANDD194 pKa = 3.92EE195 pKa = 4.73EE196 pKa = 5.12IGLAADD202 pKa = 4.02WSNGQISVAAGINLSSQGIKK222 pKa = 10.98DD223 pKa = 3.43NDD225 pKa = 3.92DD226 pKa = 3.72YY227 pKa = 11.7FIGAAYY233 pKa = 10.66NFGDD237 pKa = 3.85GNVGLNYY244 pKa = 10.11YY245 pKa = 10.25DD246 pKa = 4.31YY247 pKa = 9.38NTAAGGVSDD256 pKa = 3.9STQVTLYY263 pKa = 11.25GNYY266 pKa = 9.26TFGATTLKK274 pKa = 10.68AYY276 pKa = 10.63VSDD279 pKa = 3.83WDD281 pKa = 3.68RR282 pKa = 11.84AGYY285 pKa = 7.86DD286 pKa = 3.12TAYY289 pKa = 10.66GIGADD294 pKa = 3.68YY295 pKa = 11.25SLGEE299 pKa = 4.17GARR302 pKa = 11.84ISGSIQSGFGDD313 pKa = 3.4AAGANNGTKK322 pKa = 10.19ADD324 pKa = 3.75LGVRR328 pKa = 11.84FDD330 pKa = 3.67FF331 pKa = 5.43
Molecular weight: 34.71 kDa
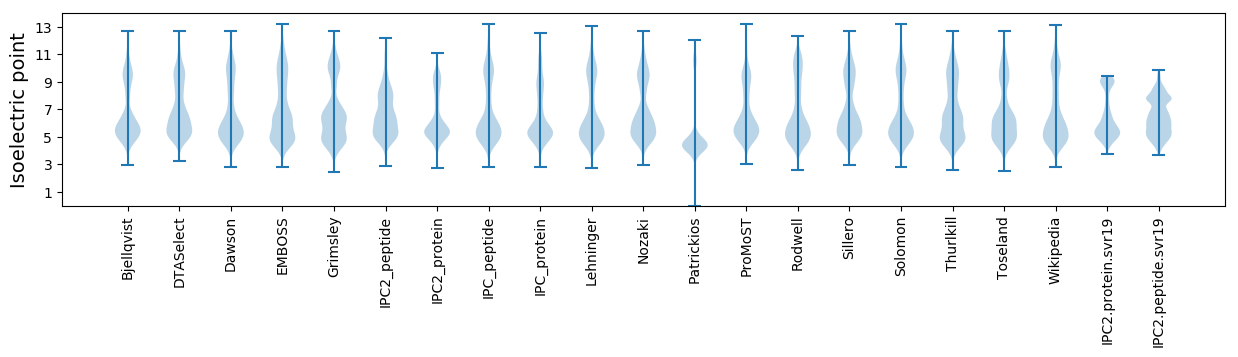
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H5AHU7|A0A1H5AHU7_9RHOB Alkyl hydroperoxide reductase C OS=Rhodobacter sp. 24-YEA-8 OX=1884310 GN=SAMN05519105_3942 PE=3 SV=1
MM1 pKa = 7.57PRR3 pKa = 11.84RR4 pKa = 11.84PRR6 pKa = 11.84VLRR9 pKa = 11.84LRR11 pKa = 11.84IGRR14 pKa = 11.84IVNLSQIVVIAILTIIIGNHH34 pKa = 4.3GG35 pKa = 3.44
MM1 pKa = 7.57PRR3 pKa = 11.84RR4 pKa = 11.84PRR6 pKa = 11.84VLRR9 pKa = 11.84LRR11 pKa = 11.84IGRR14 pKa = 11.84IVNLSQIVVIAILTIIIGNHH34 pKa = 4.3GG35 pKa = 3.44
Molecular weight: 3.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1432208 |
29 |
3632 |
316.8 |
34.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.865 ± 0.053 | 0.853 ± 0.011 |
5.563 ± 0.03 | 5.694 ± 0.027 |
3.639 ± 0.022 | 9.056 ± 0.048 |
1.958 ± 0.017 | 5.267 ± 0.025 |
2.879 ± 0.027 | 10.507 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.644 ± 0.017 | 2.419 ± 0.021 |
5.349 ± 0.031 | 3.104 ± 0.018 |
7.113 ± 0.038 | 5.321 ± 0.022 |
5.276 ± 0.022 | 6.972 ± 0.031 |
1.466 ± 0.016 | 2.056 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
