
Mastacembelus armatus (zig-zag eel)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Euteleosteomorpha; Neoteleostei; Eurypterygia;
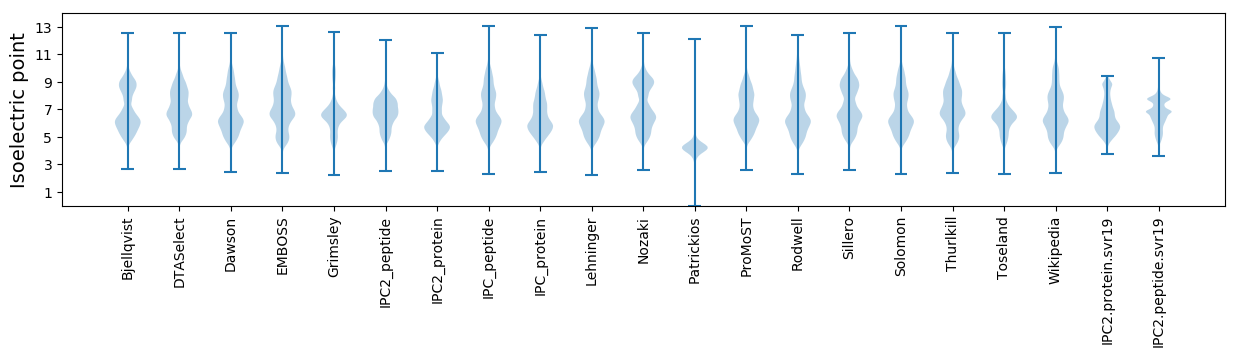
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 59116 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
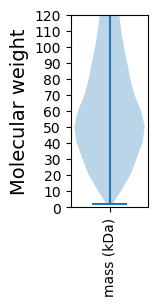
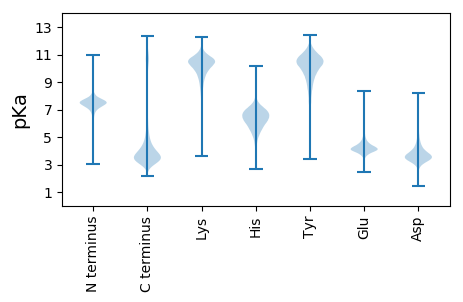
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7N8YH35|A0A7N8YH35_9TELE Isoform of A0A7N8YM48 Protein unc-13 homolog B-like OS=Mastacembelus armatus OX=205130 PE=4 SV=1
MM1 pKa = 7.42SLLSGMGGMQPRR13 pKa = 11.84PSCVPNPCHH22 pKa = 6.72PGVKK26 pKa = 9.69CVVTPEE32 pKa = 4.07GTKK35 pKa = 10.48CGPCPDD41 pKa = 3.56GTEE44 pKa = 4.51GNGTHH49 pKa = 6.04CTDD52 pKa = 2.87VDD54 pKa = 4.23EE55 pKa = 4.65VCRR58 pKa = 11.84DD59 pKa = 3.15INEE62 pKa = 4.46CEE64 pKa = 4.11GPNNGGCVEE73 pKa = 4.76NSVCMNTPGSFRR85 pKa = 11.84CGPCNPGYY93 pKa = 11.0VGDD96 pKa = 4.05QRR98 pKa = 11.84QGCKK102 pKa = 9.8PEE104 pKa = 3.97RR105 pKa = 11.84ACGNGQPNPCHH116 pKa = 6.71ASAEE120 pKa = 4.62CIVHH124 pKa = 6.66RR125 pKa = 11.84EE126 pKa = 4.01GTIEE130 pKa = 4.18CQCGVGWAGNGFLCGSDD147 pKa = 2.85IDD149 pKa = 4.15IDD151 pKa = 3.95GFPDD155 pKa = 3.4EE156 pKa = 5.89KK157 pKa = 10.96LDD159 pKa = 4.25CPEE162 pKa = 4.42RR163 pKa = 11.84NCDD166 pKa = 3.95KK167 pKa = 10.9DD168 pKa = 3.45NCVNVPNSGQEE179 pKa = 3.78DD180 pKa = 4.02ADD182 pKa = 4.12RR183 pKa = 11.84DD184 pKa = 4.5GIGDD188 pKa = 4.0ACDD191 pKa = 3.53EE192 pKa = 4.74DD193 pKa = 5.71ADD195 pKa = 4.76GDD197 pKa = 4.64GIPNTQDD204 pKa = 2.78NCVLVPNLDD213 pKa = 3.49QRR215 pKa = 11.84NVDD218 pKa = 3.55EE219 pKa = 6.13DD220 pKa = 4.52DD221 pKa = 5.62FGDD224 pKa = 3.87ACDD227 pKa = 3.29NCRR230 pKa = 11.84AVKK233 pKa = 10.88NNDD236 pKa = 3.36QKK238 pKa = 11.3DD239 pKa = 3.35TDD241 pKa = 3.93LDD243 pKa = 3.96KK244 pKa = 11.55FGDD247 pKa = 3.9EE248 pKa = 4.55CDD250 pKa = 3.68EE251 pKa = 5.81DD252 pKa = 4.18IDD254 pKa = 5.21GDD256 pKa = 4.45GIPNHH261 pKa = 7.08LDD263 pKa = 2.97NCKK266 pKa = 9.84RR267 pKa = 11.84VPNADD272 pKa = 3.2QKK274 pKa = 11.71DD275 pKa = 3.63RR276 pKa = 11.84DD277 pKa = 3.66GDD279 pKa = 4.12KK280 pKa = 11.46VGDD283 pKa = 4.05ACDD286 pKa = 3.38SCPYY290 pKa = 10.12VPNPDD295 pKa = 3.95QMDD298 pKa = 3.39VDD300 pKa = 4.01NDD302 pKa = 4.86LIGDD306 pKa = 3.96PCDD309 pKa = 3.62TNKK312 pKa = 10.93DD313 pKa = 3.49SDD315 pKa = 4.16GDD317 pKa = 3.81GHH319 pKa = 6.91QDD321 pKa = 4.2SRR323 pKa = 11.84DD324 pKa = 3.47NCPAVINSSQLDD336 pKa = 3.5TDD338 pKa = 3.78KK339 pKa = 11.37DD340 pKa = 4.07GKK342 pKa = 11.12GDD344 pKa = 3.71EE345 pKa = 5.3CDD347 pKa = 5.61DD348 pKa = 5.63DD349 pKa = 6.35DD350 pKa = 7.03DD351 pKa = 6.2NDD353 pKa = 5.44GIPDD357 pKa = 4.89LLPPGPDD364 pKa = 3.07NCRR367 pKa = 11.84LIYY370 pKa = 10.78NPLQEE375 pKa = 5.74DD376 pKa = 3.52SDD378 pKa = 4.45GDD380 pKa = 4.06GVGNVCEE387 pKa = 4.73KK388 pKa = 11.08DD389 pKa = 3.31FDD391 pKa = 4.21NDD393 pKa = 3.98TVIDD397 pKa = 4.44TIDD400 pKa = 3.4VCPEE404 pKa = 3.65NAEE407 pKa = 4.04VTLTDD412 pKa = 3.59FRR414 pKa = 11.84EE415 pKa = 4.27YY416 pKa = 9.18QTVVLDD422 pKa = 4.15PEE424 pKa = 5.32GDD426 pKa = 3.65AQIDD430 pKa = 4.13PNWVVLNQGRR440 pKa = 11.84EE441 pKa = 3.9IVQTMNSDD449 pKa = 3.19PGLAVGYY456 pKa = 7.12TAFSGVDD463 pKa = 3.72FEE465 pKa = 5.24GTFHH469 pKa = 6.81VNTVTDD475 pKa = 3.57DD476 pKa = 4.08DD477 pKa = 4.35YY478 pKa = 12.0AGFIFGYY485 pKa = 9.58QDD487 pKa = 2.83SSSFYY492 pKa = 9.91VVMWKK497 pKa = 9.52QVEE500 pKa = 4.18QIYY503 pKa = 8.33WQANPFRR510 pKa = 11.84AVAEE514 pKa = 4.08PGIQLKK520 pKa = 10.06AVKK523 pKa = 10.35SNTGPGEE530 pKa = 4.06NLRR533 pKa = 11.84NALWHH538 pKa = 6.46TGDD541 pKa = 3.25TTDD544 pKa = 4.87QVKK547 pKa = 10.41LLWKK551 pKa = 10.3DD552 pKa = 3.26SRR554 pKa = 11.84NVGWKK559 pKa = 10.42DD560 pKa = 2.97KK561 pKa = 10.83TSYY564 pKa = 10.52RR565 pKa = 11.84WFLQHH570 pKa = 6.87RR571 pKa = 11.84PADD574 pKa = 3.73GYY576 pKa = 10.45IRR578 pKa = 11.84VRR580 pKa = 11.84FYY582 pKa = 11.03EE583 pKa = 4.61GPKK586 pKa = 9.26MVADD590 pKa = 3.62TGVIIDD596 pKa = 3.35ATMRR600 pKa = 11.84GGRR603 pKa = 11.84LGVFCFSQEE612 pKa = 3.95NIIWANLRR620 pKa = 11.84YY621 pKa = 9.71RR622 pKa = 11.84CNDD625 pKa = 3.48TLPEE629 pKa = 4.41DD630 pKa = 3.9FDD632 pKa = 3.96TYY634 pKa = 10.72RR635 pKa = 11.84AQQVQLVAA643 pKa = 4.4
MM1 pKa = 7.42SLLSGMGGMQPRR13 pKa = 11.84PSCVPNPCHH22 pKa = 6.72PGVKK26 pKa = 9.69CVVTPEE32 pKa = 4.07GTKK35 pKa = 10.48CGPCPDD41 pKa = 3.56GTEE44 pKa = 4.51GNGTHH49 pKa = 6.04CTDD52 pKa = 2.87VDD54 pKa = 4.23EE55 pKa = 4.65VCRR58 pKa = 11.84DD59 pKa = 3.15INEE62 pKa = 4.46CEE64 pKa = 4.11GPNNGGCVEE73 pKa = 4.76NSVCMNTPGSFRR85 pKa = 11.84CGPCNPGYY93 pKa = 11.0VGDD96 pKa = 4.05QRR98 pKa = 11.84QGCKK102 pKa = 9.8PEE104 pKa = 3.97RR105 pKa = 11.84ACGNGQPNPCHH116 pKa = 6.71ASAEE120 pKa = 4.62CIVHH124 pKa = 6.66RR125 pKa = 11.84EE126 pKa = 4.01GTIEE130 pKa = 4.18CQCGVGWAGNGFLCGSDD147 pKa = 2.85IDD149 pKa = 4.15IDD151 pKa = 3.95GFPDD155 pKa = 3.4EE156 pKa = 5.89KK157 pKa = 10.96LDD159 pKa = 4.25CPEE162 pKa = 4.42RR163 pKa = 11.84NCDD166 pKa = 3.95KK167 pKa = 10.9DD168 pKa = 3.45NCVNVPNSGQEE179 pKa = 3.78DD180 pKa = 4.02ADD182 pKa = 4.12RR183 pKa = 11.84DD184 pKa = 4.5GIGDD188 pKa = 4.0ACDD191 pKa = 3.53EE192 pKa = 4.74DD193 pKa = 5.71ADD195 pKa = 4.76GDD197 pKa = 4.64GIPNTQDD204 pKa = 2.78NCVLVPNLDD213 pKa = 3.49QRR215 pKa = 11.84NVDD218 pKa = 3.55EE219 pKa = 6.13DD220 pKa = 4.52DD221 pKa = 5.62FGDD224 pKa = 3.87ACDD227 pKa = 3.29NCRR230 pKa = 11.84AVKK233 pKa = 10.88NNDD236 pKa = 3.36QKK238 pKa = 11.3DD239 pKa = 3.35TDD241 pKa = 3.93LDD243 pKa = 3.96KK244 pKa = 11.55FGDD247 pKa = 3.9EE248 pKa = 4.55CDD250 pKa = 3.68EE251 pKa = 5.81DD252 pKa = 4.18IDD254 pKa = 5.21GDD256 pKa = 4.45GIPNHH261 pKa = 7.08LDD263 pKa = 2.97NCKK266 pKa = 9.84RR267 pKa = 11.84VPNADD272 pKa = 3.2QKK274 pKa = 11.71DD275 pKa = 3.63RR276 pKa = 11.84DD277 pKa = 3.66GDD279 pKa = 4.12KK280 pKa = 11.46VGDD283 pKa = 4.05ACDD286 pKa = 3.38SCPYY290 pKa = 10.12VPNPDD295 pKa = 3.95QMDD298 pKa = 3.39VDD300 pKa = 4.01NDD302 pKa = 4.86LIGDD306 pKa = 3.96PCDD309 pKa = 3.62TNKK312 pKa = 10.93DD313 pKa = 3.49SDD315 pKa = 4.16GDD317 pKa = 3.81GHH319 pKa = 6.91QDD321 pKa = 4.2SRR323 pKa = 11.84DD324 pKa = 3.47NCPAVINSSQLDD336 pKa = 3.5TDD338 pKa = 3.78KK339 pKa = 11.37DD340 pKa = 4.07GKK342 pKa = 11.12GDD344 pKa = 3.71EE345 pKa = 5.3CDD347 pKa = 5.61DD348 pKa = 5.63DD349 pKa = 6.35DD350 pKa = 7.03DD351 pKa = 6.2NDD353 pKa = 5.44GIPDD357 pKa = 4.89LLPPGPDD364 pKa = 3.07NCRR367 pKa = 11.84LIYY370 pKa = 10.78NPLQEE375 pKa = 5.74DD376 pKa = 3.52SDD378 pKa = 4.45GDD380 pKa = 4.06GVGNVCEE387 pKa = 4.73KK388 pKa = 11.08DD389 pKa = 3.31FDD391 pKa = 4.21NDD393 pKa = 3.98TVIDD397 pKa = 4.44TIDD400 pKa = 3.4VCPEE404 pKa = 3.65NAEE407 pKa = 4.04VTLTDD412 pKa = 3.59FRR414 pKa = 11.84EE415 pKa = 4.27YY416 pKa = 9.18QTVVLDD422 pKa = 4.15PEE424 pKa = 5.32GDD426 pKa = 3.65AQIDD430 pKa = 4.13PNWVVLNQGRR440 pKa = 11.84EE441 pKa = 3.9IVQTMNSDD449 pKa = 3.19PGLAVGYY456 pKa = 7.12TAFSGVDD463 pKa = 3.72FEE465 pKa = 5.24GTFHH469 pKa = 6.81VNTVTDD475 pKa = 3.57DD476 pKa = 4.08DD477 pKa = 4.35YY478 pKa = 12.0AGFIFGYY485 pKa = 9.58QDD487 pKa = 2.83SSSFYY492 pKa = 9.91VVMWKK497 pKa = 9.52QVEE500 pKa = 4.18QIYY503 pKa = 8.33WQANPFRR510 pKa = 11.84AVAEE514 pKa = 4.08PGIQLKK520 pKa = 10.06AVKK523 pKa = 10.35SNTGPGEE530 pKa = 4.06NLRR533 pKa = 11.84NALWHH538 pKa = 6.46TGDD541 pKa = 3.25TTDD544 pKa = 4.87QVKK547 pKa = 10.41LLWKK551 pKa = 10.3DD552 pKa = 3.26SRR554 pKa = 11.84NVGWKK559 pKa = 10.42DD560 pKa = 2.97KK561 pKa = 10.83TSYY564 pKa = 10.52RR565 pKa = 11.84WFLQHH570 pKa = 6.87RR571 pKa = 11.84PADD574 pKa = 3.73GYY576 pKa = 10.45IRR578 pKa = 11.84VRR580 pKa = 11.84FYY582 pKa = 11.03EE583 pKa = 4.61GPKK586 pKa = 9.26MVADD590 pKa = 3.62TGVIIDD596 pKa = 3.35ATMRR600 pKa = 11.84GGRR603 pKa = 11.84LGVFCFSQEE612 pKa = 3.95NIIWANLRR620 pKa = 11.84YY621 pKa = 9.71RR622 pKa = 11.84CNDD625 pKa = 3.48TLPEE629 pKa = 4.41DD630 pKa = 3.9FDD632 pKa = 3.96TYY634 pKa = 10.72RR635 pKa = 11.84AQQVQLVAA643 pKa = 4.4
Molecular weight: 70.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q3NFI3|A0A3Q3NFI3_9TELE Uncharacterized protein OS=Mastacembelus armatus OX=205130 PE=4 SV=1
SS1 pKa = 4.63SHH3 pKa = 5.15KK4 pKa = 8.9TFRR7 pKa = 11.84IKK9 pKa = 10.64RR10 pKa = 11.84FLAKK14 pKa = 9.71KK15 pKa = 9.58QKK17 pKa = 8.69QNRR20 pKa = 11.84PIPQWIRR27 pKa = 11.84MKK29 pKa = 9.89TGNKK33 pKa = 8.61IRR35 pKa = 11.84YY36 pKa = 7.09NSKK39 pKa = 8.3RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.95WRR44 pKa = 11.84RR45 pKa = 11.84TKK47 pKa = 10.83LGLL50 pKa = 3.67
SS1 pKa = 4.63SHH3 pKa = 5.15KK4 pKa = 8.9TFRR7 pKa = 11.84IKK9 pKa = 10.64RR10 pKa = 11.84FLAKK14 pKa = 9.71KK15 pKa = 9.58QKK17 pKa = 8.69QNRR20 pKa = 11.84PIPQWIRR27 pKa = 11.84MKK29 pKa = 9.89TGNKK33 pKa = 8.61IRR35 pKa = 11.84YY36 pKa = 7.09NSKK39 pKa = 8.3RR40 pKa = 11.84RR41 pKa = 11.84HH42 pKa = 3.95WRR44 pKa = 11.84RR45 pKa = 11.84TKK47 pKa = 10.83LGLL50 pKa = 3.67
Molecular weight: 6.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
40357664 |
17 |
8413 |
682.7 |
76.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.359 ± 0.008 | 2.313 ± 0.008 |
5.24 ± 0.006 | 6.721 ± 0.01 |
3.819 ± 0.007 | 6.167 ± 0.008 |
2.669 ± 0.004 | 4.754 ± 0.007 |
5.686 ± 0.009 | 9.775 ± 0.011 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.428 ± 0.004 | 3.968 ± 0.006 |
5.376 ± 0.011 | 4.726 ± 0.008 |
5.446 ± 0.007 | 8.266 ± 0.011 |
5.669 ± 0.007 | 6.482 ± 0.008 |
1.194 ± 0.003 | 2.94 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
