
Vulpes vulpes (Red fox)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Mammalia; Theria; Eutheria;
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
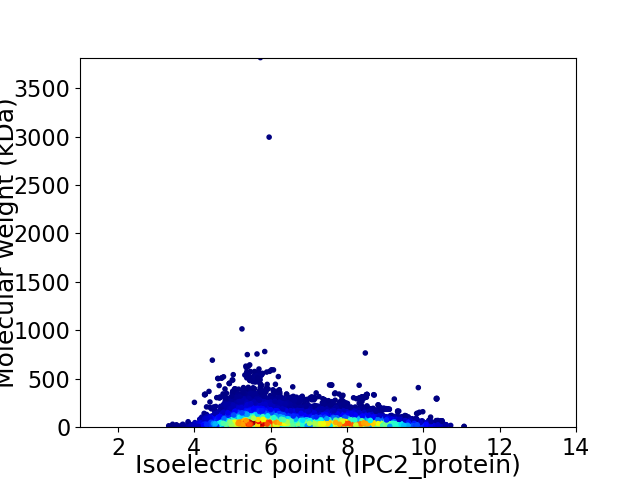
Virtual 2D-PAGE plot for 32184 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3Q7R749|A0A3Q7R749_VULVU serine/threonine-protein kinase Chk2 isoform X1 OS=Vulpes vulpes OX=9627 GN=CHEK2 PE=4 SV=1
MM1 pKa = 7.85DD2 pKa = 6.69ADD4 pKa = 4.76PEE6 pKa = 4.56TQSSCEE12 pKa = 3.95PEE14 pKa = 3.74AARR17 pKa = 11.84SVVEE21 pKa = 3.92AEE23 pKa = 3.9EE24 pKa = 5.6DD25 pKa = 3.71EE26 pKa = 4.8GCLEE30 pKa = 4.2SSHH33 pKa = 5.18TRR35 pKa = 11.84KK36 pKa = 10.0RR37 pKa = 11.84SCIPSRR43 pKa = 11.84GSWHH47 pKa = 6.91LLLPQALLQLLTDD60 pKa = 4.87PAPAPDD66 pKa = 4.35PALAPDD72 pKa = 5.17PDD74 pKa = 4.65PAPDD78 pKa = 4.2PAPAPDD84 pKa = 4.43SVPAPDD90 pKa = 4.41SAPAPDD96 pKa = 4.18PAAALASATTSVPCPHH112 pKa = 6.58TTPPPAPAPAPAPAAASDD130 pKa = 3.79PAPARR135 pKa = 11.84EE136 pKa = 4.09TAPDD140 pKa = 3.63PAAAPVPAPTPATASCPEE158 pKa = 4.08STPASNTDD166 pKa = 3.85PVPSPAPDD174 pKa = 3.98PSPDD178 pKa = 4.08CDD180 pKa = 4.22HH181 pKa = 7.52DD182 pKa = 4.16PAPASAPDD190 pKa = 3.87PALTSAPDD198 pKa = 3.92DD199 pKa = 4.58DD200 pKa = 4.84LASAAVPAPALNTEE214 pKa = 4.96FGPDD218 pKa = 3.72PVPAPDD224 pKa = 4.9LEE226 pKa = 5.63NDD228 pKa = 4.46LSPAPAPAAAPAGAPAPDD246 pKa = 3.46TDD248 pKa = 4.6FPFDD252 pKa = 3.79SAPDD256 pKa = 3.75TEE258 pKa = 4.95LAPACTPDD266 pKa = 3.68PNLAHH271 pKa = 6.52VPAPVPVPDD280 pKa = 3.82TASANTPAPDD290 pKa = 3.55NDD292 pKa = 4.26LAPSPSPDD300 pKa = 3.65PAPVCDD306 pKa = 4.43TDD308 pKa = 4.41CSSASAPDD316 pKa = 3.84NEE318 pKa = 4.7PASTPALDD326 pKa = 4.61LAPHH330 pKa = 6.87TDD332 pKa = 3.86PAPTPAPAPTCAPGTDD348 pKa = 4.06TDD350 pKa = 4.6PVPAPDD356 pKa = 4.17NDD358 pKa = 4.36LAPGPAPDD366 pKa = 3.9TAPGPVIAPAPGLALVPTPGLAPPPDD392 pKa = 3.83NDD394 pKa = 3.43FASAPGPDD402 pKa = 4.75MYY404 pKa = 10.65PAPAPGLDD412 pKa = 4.07RR413 pKa = 11.84DD414 pKa = 4.24PTPAPAPPPVPAGLPEE430 pKa = 4.13THH432 pKa = 7.16LEE434 pKa = 3.99PTALRR439 pKa = 11.84GDD441 pKa = 4.61LLVQSPFSPSPDD453 pKa = 3.77PDD455 pKa = 4.09PLPTCHH461 pKa = 7.43PLPLWSFFLVSVHH474 pKa = 6.21CLSIFTHH481 pKa = 6.6SDD483 pKa = 3.12LFQKK487 pKa = 9.22VVPYY491 pKa = 10.71SCLGYY496 pKa = 9.61MRR498 pKa = 11.84SQQVKK503 pKa = 9.78KK504 pKa = 10.7SNEE507 pKa = 3.87HH508 pKa = 6.32LPPTVHH514 pKa = 7.67AIITT518 pKa = 3.75
MM1 pKa = 7.85DD2 pKa = 6.69ADD4 pKa = 4.76PEE6 pKa = 4.56TQSSCEE12 pKa = 3.95PEE14 pKa = 3.74AARR17 pKa = 11.84SVVEE21 pKa = 3.92AEE23 pKa = 3.9EE24 pKa = 5.6DD25 pKa = 3.71EE26 pKa = 4.8GCLEE30 pKa = 4.2SSHH33 pKa = 5.18TRR35 pKa = 11.84KK36 pKa = 10.0RR37 pKa = 11.84SCIPSRR43 pKa = 11.84GSWHH47 pKa = 6.91LLLPQALLQLLTDD60 pKa = 4.87PAPAPDD66 pKa = 4.35PALAPDD72 pKa = 5.17PDD74 pKa = 4.65PAPDD78 pKa = 4.2PAPAPDD84 pKa = 4.43SVPAPDD90 pKa = 4.41SAPAPDD96 pKa = 4.18PAAALASATTSVPCPHH112 pKa = 6.58TTPPPAPAPAPAPAAASDD130 pKa = 3.79PAPARR135 pKa = 11.84EE136 pKa = 4.09TAPDD140 pKa = 3.63PAAAPVPAPTPATASCPEE158 pKa = 4.08STPASNTDD166 pKa = 3.85PVPSPAPDD174 pKa = 3.98PSPDD178 pKa = 4.08CDD180 pKa = 4.22HH181 pKa = 7.52DD182 pKa = 4.16PAPASAPDD190 pKa = 3.87PALTSAPDD198 pKa = 3.92DD199 pKa = 4.58DD200 pKa = 4.84LASAAVPAPALNTEE214 pKa = 4.96FGPDD218 pKa = 3.72PVPAPDD224 pKa = 4.9LEE226 pKa = 5.63NDD228 pKa = 4.46LSPAPAPAAAPAGAPAPDD246 pKa = 3.46TDD248 pKa = 4.6FPFDD252 pKa = 3.79SAPDD256 pKa = 3.75TEE258 pKa = 4.95LAPACTPDD266 pKa = 3.68PNLAHH271 pKa = 6.52VPAPVPVPDD280 pKa = 3.82TASANTPAPDD290 pKa = 3.55NDD292 pKa = 4.26LAPSPSPDD300 pKa = 3.65PAPVCDD306 pKa = 4.43TDD308 pKa = 4.41CSSASAPDD316 pKa = 3.84NEE318 pKa = 4.7PASTPALDD326 pKa = 4.61LAPHH330 pKa = 6.87TDD332 pKa = 3.86PAPTPAPAPTCAPGTDD348 pKa = 4.06TDD350 pKa = 4.6PVPAPDD356 pKa = 4.17NDD358 pKa = 4.36LAPGPAPDD366 pKa = 3.9TAPGPVIAPAPGLALVPTPGLAPPPDD392 pKa = 3.83NDD394 pKa = 3.43FASAPGPDD402 pKa = 4.75MYY404 pKa = 10.65PAPAPGLDD412 pKa = 4.07RR413 pKa = 11.84DD414 pKa = 4.24PTPAPAPPPVPAGLPEE430 pKa = 4.13THH432 pKa = 7.16LEE434 pKa = 3.99PTALRR439 pKa = 11.84GDD441 pKa = 4.61LLVQSPFSPSPDD453 pKa = 3.77PDD455 pKa = 4.09PLPTCHH461 pKa = 7.43PLPLWSFFLVSVHH474 pKa = 6.21CLSIFTHH481 pKa = 6.6SDD483 pKa = 3.12LFQKK487 pKa = 9.22VVPYY491 pKa = 10.71SCLGYY496 pKa = 9.61MRR498 pKa = 11.84SQQVKK503 pKa = 9.78KK504 pKa = 10.7SNEE507 pKa = 3.87HH508 pKa = 6.32LPPTVHH514 pKa = 7.67AIITT518 pKa = 3.75
Molecular weight: 51.77 kDa
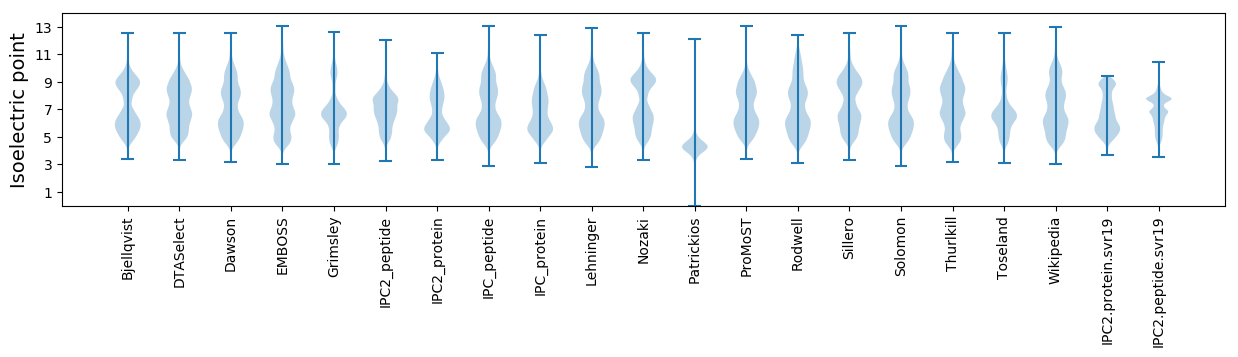
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3Q7UJM5|A0A3Q7UJM5_VULVU GTPase IMAP family member GIMD1 OS=Vulpes vulpes OX=9627 GN=GIMD1 PE=3 SV=1
PP1 pKa = 7.05HH2 pKa = 7.7RR3 pKa = 11.84APGAWSLLLPLPMSMSSHH21 pKa = 4.44KK22 pKa = 8.94TFRR25 pKa = 11.84IKK27 pKa = 10.64RR28 pKa = 11.84FLAKK32 pKa = 9.71KK33 pKa = 9.58QKK35 pKa = 8.69QNRR38 pKa = 11.84PIPQWIQMKK47 pKa = 8.88TGNKK51 pKa = 8.6IRR53 pKa = 11.84YY54 pKa = 7.09NSKK57 pKa = 8.3RR58 pKa = 11.84RR59 pKa = 11.84HH60 pKa = 3.95WRR62 pKa = 11.84RR63 pKa = 11.84TKK65 pKa = 10.83LGLL68 pKa = 3.67
PP1 pKa = 7.05HH2 pKa = 7.7RR3 pKa = 11.84APGAWSLLLPLPMSMSSHH21 pKa = 4.44KK22 pKa = 8.94TFRR25 pKa = 11.84IKK27 pKa = 10.64RR28 pKa = 11.84FLAKK32 pKa = 9.71KK33 pKa = 9.58QKK35 pKa = 8.69QNRR38 pKa = 11.84PIPQWIQMKK47 pKa = 8.88TGNKK51 pKa = 8.6IRR53 pKa = 11.84YY54 pKa = 7.09NSKK57 pKa = 8.3RR58 pKa = 11.84RR59 pKa = 11.84HH60 pKa = 3.95WRR62 pKa = 11.84RR63 pKa = 11.84TKK65 pKa = 10.83LGLL68 pKa = 3.67
Molecular weight: 8.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
21246518 |
15 |
34362 |
660.2 |
73.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.955 ± 0.015 | 2.115 ± 0.01 |
4.857 ± 0.009 | 7.257 ± 0.019 |
3.497 ± 0.008 | 6.482 ± 0.019 |
2.572 ± 0.007 | 4.317 ± 0.013 |
5.8 ± 0.019 | 9.806 ± 0.02 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.147 ± 0.006 | 3.583 ± 0.009 |
6.47 ± 0.025 | 4.875 ± 0.014 |
5.736 ± 0.013 | 8.516 ± 0.02 |
5.316 ± 0.013 | 5.952 ± 0.014 |
1.139 ± 0.004 | 2.567 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
