
Gayfeather mild mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Cucumovirus
Average proteome isoelectric point is 6.73
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|C0MNB4|C0MNB4_9BROM 2b protein OS=Gayfeather mild mottle virus OX=578305 GN=2b PE=4 SV=1
MM1 pKa = 7.83ASTDD5 pKa = 3.68VIFSLEE11 pKa = 3.77NLLNGSYY18 pKa = 10.96GVVTPEE24 pKa = 3.98DD25 pKa = 3.52DD26 pKa = 3.47ALFRR30 pKa = 11.84QRR32 pKa = 11.84SMYY35 pKa = 9.89DD36 pKa = 2.94ASVSDD41 pKa = 4.86DD42 pKa = 4.81SIITDD47 pKa = 4.35DD48 pKa = 4.38DD49 pKa = 3.98DD50 pKa = 5.68AVAFTCDD57 pKa = 3.41DD58 pKa = 3.41VSADD62 pKa = 3.35GHH64 pKa = 6.14GEE66 pKa = 4.25SNPSDD71 pKa = 3.65PCSEE75 pKa = 5.16DD76 pKa = 3.07GCQFNDD82 pKa = 3.35NPVLYY87 pKa = 10.45DD88 pKa = 3.31IEE90 pKa = 4.78EE91 pKa = 4.2YY92 pKa = 10.42QALSDD97 pKa = 4.13SEE99 pKa = 5.0LFALFDD105 pKa = 4.57TIVKK109 pKa = 9.27PIKK112 pKa = 9.98FGSILTPEE120 pKa = 4.65FSRR123 pKa = 11.84PRR125 pKa = 11.84FYY127 pKa = 10.65TALSLAKK134 pKa = 9.78SVSVISNRR142 pKa = 11.84PSTFGEE148 pKa = 4.2VSPEE152 pKa = 4.16LIKK155 pKa = 10.71AIYY158 pKa = 9.28IGEE161 pKa = 5.04PITSVSGAHH170 pKa = 6.37AVGDD174 pKa = 4.04TPDD177 pKa = 3.66DD178 pKa = 3.74CEE180 pKa = 5.44GYY182 pKa = 10.68VFVPSEE188 pKa = 3.94PTSNFEE194 pKa = 4.05PPPICEE200 pKa = 4.14EE201 pKa = 4.25CGLAAYY207 pKa = 8.06HH208 pKa = 6.64CPHH211 pKa = 7.0FDD213 pKa = 3.48FNALKK218 pKa = 8.62EE219 pKa = 4.26THH221 pKa = 6.49NDD223 pKa = 2.81HH224 pKa = 6.79TISHH228 pKa = 7.32DD229 pKa = 3.88YY230 pKa = 10.63EE231 pKa = 5.6IEE233 pKa = 4.12TLTGVIDD240 pKa = 5.07DD241 pKa = 4.49ATLLMNLGPFLVPIQCEE258 pKa = 3.83YY259 pKa = 11.57SKK261 pKa = 9.93TFEE264 pKa = 4.59SKK266 pKa = 10.52HH267 pKa = 4.5ATKK270 pKa = 10.31PSLARR275 pKa = 11.84PTDD278 pKa = 3.52RR279 pKa = 11.84VDD281 pKa = 3.14VDD283 pKa = 3.75VVQAVCDD290 pKa = 3.92SMLPTHH296 pKa = 6.68VNYY299 pKa = 10.61DD300 pKa = 3.53DD301 pKa = 4.66TYY303 pKa = 9.07HH304 pKa = 5.8QVFVEE309 pKa = 4.29DD310 pKa = 3.31SDD312 pKa = 4.5YY313 pKa = 11.69SVDD316 pKa = 3.29IDD318 pKa = 5.17RR319 pKa = 11.84IRR321 pKa = 11.84LKK323 pKa = 10.79QSDD326 pKa = 4.85LLPKK330 pKa = 10.52VSDD333 pKa = 4.56DD334 pKa = 3.5GHH336 pKa = 5.5MHH338 pKa = 6.63PVLNTGSGHH347 pKa = 6.19KK348 pKa = 10.0RR349 pKa = 11.84VGTQKK354 pKa = 10.21EE355 pKa = 3.95VHH357 pKa = 5.29TAIKK361 pKa = 9.92KK362 pKa = 9.94RR363 pKa = 11.84NADD366 pKa = 3.49VPEE369 pKa = 5.43LGDD372 pKa = 3.84SVNLTRR378 pKa = 11.84LSEE381 pKa = 4.14AVAEE385 pKa = 4.37RR386 pKa = 11.84FISSYY391 pKa = 10.49MNVNALVSSNFINVVGNFHH410 pKa = 7.75AYY412 pKa = 5.51MQKK415 pKa = 8.24WHH417 pKa = 6.91SSLSYY422 pKa = 11.27DD423 pKa = 5.43DD424 pKa = 6.08LPDD427 pKa = 4.0LNAEE431 pKa = 3.83NLQFYY436 pKa = 9.9EE437 pKa = 4.57HH438 pKa = 6.27MVKK441 pKa = 10.41SDD443 pKa = 3.47VKK445 pKa = 10.83PSVTDD450 pKa = 3.2TLNVDD455 pKa = 3.69RR456 pKa = 11.84PVPATITFHH465 pKa = 6.94KK466 pKa = 10.27KK467 pKa = 10.11QITSQFSPLFTALFEE482 pKa = 4.23RR483 pKa = 11.84FQRR486 pKa = 11.84CLRR489 pKa = 11.84SKK491 pKa = 10.77VVLPVGKK498 pKa = 9.93ISSLEE503 pKa = 3.97LEE505 pKa = 4.67DD506 pKa = 4.67FSVIGKK512 pKa = 8.83HH513 pKa = 5.79CLEE516 pKa = 4.76IDD518 pKa = 3.42LSKK521 pKa = 10.66FDD523 pKa = 3.77KK524 pKa = 11.17SQGEE528 pKa = 4.26LHH530 pKa = 7.24LMIQEE535 pKa = 4.23QILNRR540 pKa = 11.84LGCPAHH546 pKa = 6.52VSKK549 pKa = 9.66WWCDD553 pKa = 3.94FHH555 pKa = 7.13RR556 pKa = 11.84QSYY559 pKa = 10.55IKK561 pKa = 10.22DD562 pKa = 3.05KK563 pKa = 10.76RR564 pKa = 11.84AGVGMALSFQRR575 pKa = 11.84RR576 pKa = 11.84TGDD579 pKa = 2.78AFTYY583 pKa = 10.33FGNTLVTMALFCWCYY598 pKa = 10.03DD599 pKa = 3.43TEE601 pKa = 4.39QFDD604 pKa = 4.07RR605 pKa = 11.84MLFSGDD611 pKa = 3.53DD612 pKa = 3.49SLAFSSIAPVGDD624 pKa = 3.37PSKK627 pKa = 10.44FTTLFNMEE635 pKa = 4.51AKK637 pKa = 10.46VMEE640 pKa = 4.39PSVPYY645 pKa = 10.05ICSKK649 pKa = 10.76FLLTDD654 pKa = 3.4EE655 pKa = 5.17FGQTFSVPDD664 pKa = 4.32PLRR667 pKa = 11.84EE668 pKa = 3.88IQRR671 pKa = 11.84LGSKK675 pKa = 10.01KK676 pKa = 9.99IPYY679 pKa = 9.92SDD681 pKa = 4.79DD682 pKa = 4.37DD683 pKa = 4.53SFLHH687 pKa = 5.43AHH689 pKa = 6.21FMSFVDD695 pKa = 3.25RR696 pKa = 11.84LKK698 pKa = 10.69FLEE701 pKa = 4.45RR702 pKa = 11.84MSQFTIDD709 pKa = 3.66QLTLFYY715 pKa = 10.93EE716 pKa = 4.45MKK718 pKa = 9.82YY719 pKa = 10.41KK720 pKa = 10.73KK721 pKa = 10.34SGADD725 pKa = 3.08ASLVLGAFKK734 pKa = 10.79KK735 pKa = 9.65YY736 pKa = 5.61TTNFNAYY743 pKa = 9.19KK744 pKa = 10.07EE745 pKa = 5.0LYY747 pKa = 10.17FSDD750 pKa = 4.16RR751 pKa = 11.84HH752 pKa = 5.3QCDD755 pKa = 4.47LINSFSIGDD764 pKa = 3.84FVIKK768 pKa = 10.48LYY770 pKa = 10.14VEE772 pKa = 4.3KK773 pKa = 10.08PARR776 pKa = 11.84RR777 pKa = 3.59
MM1 pKa = 7.83ASTDD5 pKa = 3.68VIFSLEE11 pKa = 3.77NLLNGSYY18 pKa = 10.96GVVTPEE24 pKa = 3.98DD25 pKa = 3.52DD26 pKa = 3.47ALFRR30 pKa = 11.84QRR32 pKa = 11.84SMYY35 pKa = 9.89DD36 pKa = 2.94ASVSDD41 pKa = 4.86DD42 pKa = 4.81SIITDD47 pKa = 4.35DD48 pKa = 4.38DD49 pKa = 3.98DD50 pKa = 5.68AVAFTCDD57 pKa = 3.41DD58 pKa = 3.41VSADD62 pKa = 3.35GHH64 pKa = 6.14GEE66 pKa = 4.25SNPSDD71 pKa = 3.65PCSEE75 pKa = 5.16DD76 pKa = 3.07GCQFNDD82 pKa = 3.35NPVLYY87 pKa = 10.45DD88 pKa = 3.31IEE90 pKa = 4.78EE91 pKa = 4.2YY92 pKa = 10.42QALSDD97 pKa = 4.13SEE99 pKa = 5.0LFALFDD105 pKa = 4.57TIVKK109 pKa = 9.27PIKK112 pKa = 9.98FGSILTPEE120 pKa = 4.65FSRR123 pKa = 11.84PRR125 pKa = 11.84FYY127 pKa = 10.65TALSLAKK134 pKa = 9.78SVSVISNRR142 pKa = 11.84PSTFGEE148 pKa = 4.2VSPEE152 pKa = 4.16LIKK155 pKa = 10.71AIYY158 pKa = 9.28IGEE161 pKa = 5.04PITSVSGAHH170 pKa = 6.37AVGDD174 pKa = 4.04TPDD177 pKa = 3.66DD178 pKa = 3.74CEE180 pKa = 5.44GYY182 pKa = 10.68VFVPSEE188 pKa = 3.94PTSNFEE194 pKa = 4.05PPPICEE200 pKa = 4.14EE201 pKa = 4.25CGLAAYY207 pKa = 8.06HH208 pKa = 6.64CPHH211 pKa = 7.0FDD213 pKa = 3.48FNALKK218 pKa = 8.62EE219 pKa = 4.26THH221 pKa = 6.49NDD223 pKa = 2.81HH224 pKa = 6.79TISHH228 pKa = 7.32DD229 pKa = 3.88YY230 pKa = 10.63EE231 pKa = 5.6IEE233 pKa = 4.12TLTGVIDD240 pKa = 5.07DD241 pKa = 4.49ATLLMNLGPFLVPIQCEE258 pKa = 3.83YY259 pKa = 11.57SKK261 pKa = 9.93TFEE264 pKa = 4.59SKK266 pKa = 10.52HH267 pKa = 4.5ATKK270 pKa = 10.31PSLARR275 pKa = 11.84PTDD278 pKa = 3.52RR279 pKa = 11.84VDD281 pKa = 3.14VDD283 pKa = 3.75VVQAVCDD290 pKa = 3.92SMLPTHH296 pKa = 6.68VNYY299 pKa = 10.61DD300 pKa = 3.53DD301 pKa = 4.66TYY303 pKa = 9.07HH304 pKa = 5.8QVFVEE309 pKa = 4.29DD310 pKa = 3.31SDD312 pKa = 4.5YY313 pKa = 11.69SVDD316 pKa = 3.29IDD318 pKa = 5.17RR319 pKa = 11.84IRR321 pKa = 11.84LKK323 pKa = 10.79QSDD326 pKa = 4.85LLPKK330 pKa = 10.52VSDD333 pKa = 4.56DD334 pKa = 3.5GHH336 pKa = 5.5MHH338 pKa = 6.63PVLNTGSGHH347 pKa = 6.19KK348 pKa = 10.0RR349 pKa = 11.84VGTQKK354 pKa = 10.21EE355 pKa = 3.95VHH357 pKa = 5.29TAIKK361 pKa = 9.92KK362 pKa = 9.94RR363 pKa = 11.84NADD366 pKa = 3.49VPEE369 pKa = 5.43LGDD372 pKa = 3.84SVNLTRR378 pKa = 11.84LSEE381 pKa = 4.14AVAEE385 pKa = 4.37RR386 pKa = 11.84FISSYY391 pKa = 10.49MNVNALVSSNFINVVGNFHH410 pKa = 7.75AYY412 pKa = 5.51MQKK415 pKa = 8.24WHH417 pKa = 6.91SSLSYY422 pKa = 11.27DD423 pKa = 5.43DD424 pKa = 6.08LPDD427 pKa = 4.0LNAEE431 pKa = 3.83NLQFYY436 pKa = 9.9EE437 pKa = 4.57HH438 pKa = 6.27MVKK441 pKa = 10.41SDD443 pKa = 3.47VKK445 pKa = 10.83PSVTDD450 pKa = 3.2TLNVDD455 pKa = 3.69RR456 pKa = 11.84PVPATITFHH465 pKa = 6.94KK466 pKa = 10.27KK467 pKa = 10.11QITSQFSPLFTALFEE482 pKa = 4.23RR483 pKa = 11.84FQRR486 pKa = 11.84CLRR489 pKa = 11.84SKK491 pKa = 10.77VVLPVGKK498 pKa = 9.93ISSLEE503 pKa = 3.97LEE505 pKa = 4.67DD506 pKa = 4.67FSVIGKK512 pKa = 8.83HH513 pKa = 5.79CLEE516 pKa = 4.76IDD518 pKa = 3.42LSKK521 pKa = 10.66FDD523 pKa = 3.77KK524 pKa = 11.17SQGEE528 pKa = 4.26LHH530 pKa = 7.24LMIQEE535 pKa = 4.23QILNRR540 pKa = 11.84LGCPAHH546 pKa = 6.52VSKK549 pKa = 9.66WWCDD553 pKa = 3.94FHH555 pKa = 7.13RR556 pKa = 11.84QSYY559 pKa = 10.55IKK561 pKa = 10.22DD562 pKa = 3.05KK563 pKa = 10.76RR564 pKa = 11.84AGVGMALSFQRR575 pKa = 11.84RR576 pKa = 11.84TGDD579 pKa = 2.78AFTYY583 pKa = 10.33FGNTLVTMALFCWCYY598 pKa = 10.03DD599 pKa = 3.43TEE601 pKa = 4.39QFDD604 pKa = 4.07RR605 pKa = 11.84MLFSGDD611 pKa = 3.53DD612 pKa = 3.49SLAFSSIAPVGDD624 pKa = 3.37PSKK627 pKa = 10.44FTTLFNMEE635 pKa = 4.51AKK637 pKa = 10.46VMEE640 pKa = 4.39PSVPYY645 pKa = 10.05ICSKK649 pKa = 10.76FLLTDD654 pKa = 3.4EE655 pKa = 5.17FGQTFSVPDD664 pKa = 4.32PLRR667 pKa = 11.84EE668 pKa = 3.88IQRR671 pKa = 11.84LGSKK675 pKa = 10.01KK676 pKa = 9.99IPYY679 pKa = 9.92SDD681 pKa = 4.79DD682 pKa = 4.37DD683 pKa = 4.53SFLHH687 pKa = 5.43AHH689 pKa = 6.21FMSFVDD695 pKa = 3.25RR696 pKa = 11.84LKK698 pKa = 10.69FLEE701 pKa = 4.45RR702 pKa = 11.84MSQFTIDD709 pKa = 3.66QLTLFYY715 pKa = 10.93EE716 pKa = 4.45MKK718 pKa = 9.82YY719 pKa = 10.41KK720 pKa = 10.73KK721 pKa = 10.34SGADD725 pKa = 3.08ASLVLGAFKK734 pKa = 10.79KK735 pKa = 9.65YY736 pKa = 5.61TTNFNAYY743 pKa = 9.19KK744 pKa = 10.07EE745 pKa = 5.0LYY747 pKa = 10.17FSDD750 pKa = 4.16RR751 pKa = 11.84HH752 pKa = 5.3QCDD755 pKa = 4.47LINSFSIGDD764 pKa = 3.84FVIKK768 pKa = 10.48LYY770 pKa = 10.14VEE772 pKa = 4.3KK773 pKa = 10.08PARR776 pKa = 11.84RR777 pKa = 3.59
Molecular weight: 87.55 kDa
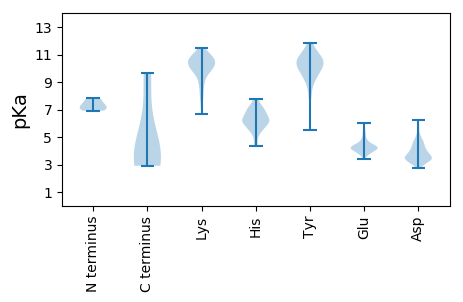
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0MNB6|C0MNB6_9BROM Capsid protein OS=Gayfeather mild mottle virus OX=578305 GN=cp PE=3 SV=1
MM1 pKa = 7.18AQSGTGGSSRR11 pKa = 11.84RR12 pKa = 11.84PRR14 pKa = 11.84RR15 pKa = 11.84GRR17 pKa = 11.84RR18 pKa = 11.84NNNNRR23 pKa = 11.84NNDD26 pKa = 3.21RR27 pKa = 11.84DD28 pKa = 3.56KK29 pKa = 11.35ALRR32 pKa = 11.84DD33 pKa = 3.13LTVQVKK39 pKa = 9.96RR40 pKa = 11.84LAIIAASTAPSLQHH54 pKa = 5.5PTFIASQRR62 pKa = 11.84CRR64 pKa = 11.84PGYY67 pKa = 9.3TYY69 pKa = 10.71TSLDD73 pKa = 3.28VRR75 pKa = 11.84PTKK78 pKa = 9.23TEE80 pKa = 3.71KK81 pKa = 10.59GHH83 pKa = 6.88SFGQRR88 pKa = 11.84LTIPVPVSDD97 pKa = 4.09YY98 pKa = 10.35PKK100 pKa = 10.6KK101 pKa = 10.39KK102 pKa = 9.77VSCVQVRR109 pKa = 11.84LNPSPKK115 pKa = 9.1FDD117 pKa = 3.39STIWVSLRR125 pKa = 11.84RR126 pKa = 11.84LDD128 pKa = 3.37EE129 pKa = 4.21TTILTSEE136 pKa = 4.2NVFKK140 pKa = 11.3LFTDD144 pKa = 5.01GAASVLIYY152 pKa = 9.41QHH154 pKa = 5.69VTTGVQPNNKK164 pKa = 8.2ITFDD168 pKa = 3.6MSNVDD173 pKa = 4.76AEE175 pKa = 4.21IGDD178 pKa = 3.83MNRR181 pKa = 11.84YY182 pKa = 9.3ALIVYY187 pKa = 10.31SKK189 pKa = 11.29DD190 pKa = 3.38DD191 pKa = 3.49VLEE194 pKa = 4.46ADD196 pKa = 3.76EE197 pKa = 4.52MVIHH201 pKa = 6.73VDD203 pKa = 3.51VEE205 pKa = 4.22HH206 pKa = 6.35QRR208 pKa = 11.84IPSATSLPVV217 pKa = 2.91
MM1 pKa = 7.18AQSGTGGSSRR11 pKa = 11.84RR12 pKa = 11.84PRR14 pKa = 11.84RR15 pKa = 11.84GRR17 pKa = 11.84RR18 pKa = 11.84NNNNRR23 pKa = 11.84NNDD26 pKa = 3.21RR27 pKa = 11.84DD28 pKa = 3.56KK29 pKa = 11.35ALRR32 pKa = 11.84DD33 pKa = 3.13LTVQVKK39 pKa = 9.96RR40 pKa = 11.84LAIIAASTAPSLQHH54 pKa = 5.5PTFIASQRR62 pKa = 11.84CRR64 pKa = 11.84PGYY67 pKa = 9.3TYY69 pKa = 10.71TSLDD73 pKa = 3.28VRR75 pKa = 11.84PTKK78 pKa = 9.23TEE80 pKa = 3.71KK81 pKa = 10.59GHH83 pKa = 6.88SFGQRR88 pKa = 11.84LTIPVPVSDD97 pKa = 4.09YY98 pKa = 10.35PKK100 pKa = 10.6KK101 pKa = 10.39KK102 pKa = 9.77VSCVQVRR109 pKa = 11.84LNPSPKK115 pKa = 9.1FDD117 pKa = 3.39STIWVSLRR125 pKa = 11.84RR126 pKa = 11.84LDD128 pKa = 3.37EE129 pKa = 4.21TTILTSEE136 pKa = 4.2NVFKK140 pKa = 11.3LFTDD144 pKa = 5.01GAASVLIYY152 pKa = 9.41QHH154 pKa = 5.69VTTGVQPNNKK164 pKa = 8.2ITFDD168 pKa = 3.6MSNVDD173 pKa = 4.76AEE175 pKa = 4.21IGDD178 pKa = 3.83MNRR181 pKa = 11.84YY182 pKa = 9.3ALIVYY187 pKa = 10.31SKK189 pKa = 11.29DD190 pKa = 3.38DD191 pKa = 3.49VLEE194 pKa = 4.46ADD196 pKa = 3.76EE197 pKa = 4.52MVIHH201 pKa = 6.73VDD203 pKa = 3.51VEE205 pKa = 4.22HH206 pKa = 6.35QRR208 pKa = 11.84IPSATSLPVV217 pKa = 2.91
Molecular weight: 24.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2382 |
120 |
988 |
476.4 |
53.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.549 ± 0.574 | 2.141 ± 0.153 |
7.095 ± 0.861 | 5.5 ± 0.326 |
5.122 ± 0.711 | 5.248 ± 0.444 |
3.107 ± 0.227 | 4.492 ± 0.396 |
6.003 ± 0.349 | 8.564 ± 0.402 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.519 ± 0.312 | 4.072 ± 0.219 |
4.492 ± 0.752 | 3.568 ± 0.191 |
5.206 ± 0.691 | 8.606 ± 0.925 |
6.549 ± 0.707 | 7.263 ± 0.281 |
0.924 ± 0.177 | 2.981 ± 0.257 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
