
Rhizoctonia fumigata mycovirus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
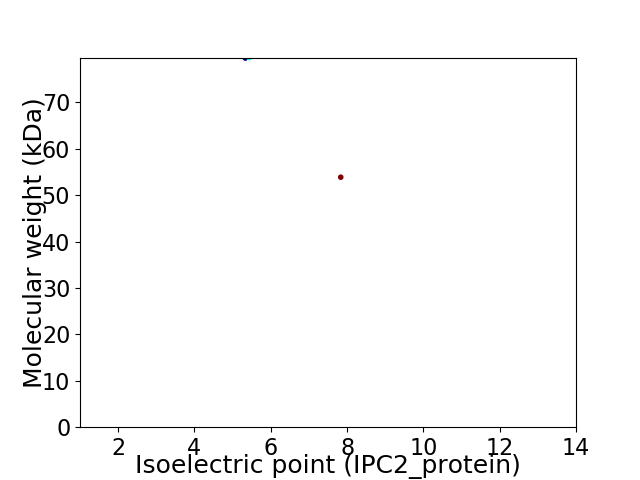
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F6Q4A5|A0A0F6Q4A5_9VIRU Putative RNA-dependent RNA polymerase OS=Rhizoctonia fumigata mycovirus OX=1592828 PE=4 SV=1
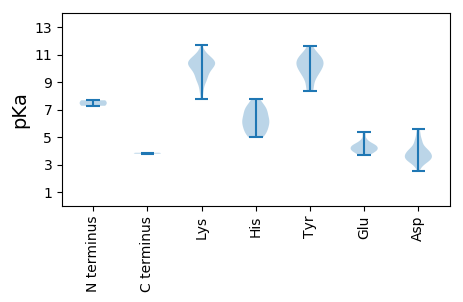
MM1 pKa = 7.67DD2 pKa = 4.7HH3 pKa = 7.13ARR5 pKa = 11.84DD6 pKa = 3.49FDD8 pKa = 4.73VISSNATNPLKK19 pKa = 10.58RR20 pKa = 11.84SRR22 pKa = 11.84EE23 pKa = 3.94EE24 pKa = 3.98ADD26 pKa = 3.39LGTAGEE32 pKa = 4.5APWRR36 pKa = 11.84TPVNGATINGVFRR49 pKa = 11.84IRR51 pKa = 11.84KK52 pKa = 9.16FDD54 pKa = 3.38VDD56 pKa = 4.0YY57 pKa = 11.63LNGPVADD64 pKa = 4.21AEE66 pKa = 4.58VVIAGLPRR74 pKa = 11.84VKK76 pKa = 10.31EE77 pKa = 4.15VVPEE81 pKa = 4.12SCDD84 pKa = 3.11ALRR87 pKa = 11.84GMKK90 pKa = 9.74VLNDD94 pKa = 3.64ADD96 pKa = 4.86HH97 pKa = 6.5ITTAVSSEE105 pKa = 4.22SLSGTGNGGTRR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84NSRR121 pKa = 11.84RR122 pKa = 11.84SRR124 pKa = 11.84KK125 pKa = 9.9NKK127 pKa = 9.7VKK129 pKa = 8.44TTAKK133 pKa = 10.37SGGSLDD139 pKa = 4.27DD140 pKa = 3.86LKK142 pKa = 11.68EE143 pKa = 4.07MFKK146 pKa = 10.76HH147 pKa = 5.1ITVHH151 pKa = 6.61DD152 pKa = 4.01AQVRR156 pKa = 11.84LGKK159 pKa = 10.24FHH161 pKa = 7.8DD162 pKa = 5.16EE163 pKa = 4.06ITFTPADD170 pKa = 3.95PAVLGYY176 pKa = 9.93LQSPEE181 pKa = 4.05GEE183 pKa = 4.01IPGFDD188 pKa = 3.33PSEE191 pKa = 4.22YY192 pKa = 10.69CFMEE196 pKa = 4.4ASGGVQMRR204 pKa = 11.84HH205 pKa = 5.06LKK207 pKa = 9.94MYY209 pKa = 10.92DD210 pKa = 3.35EE211 pKa = 4.93DD212 pKa = 4.22TSLGPDD218 pKa = 4.05FSLLEE223 pKa = 4.42DD224 pKa = 4.02PEE226 pKa = 4.75AGRR229 pKa = 11.84RR230 pKa = 11.84ALEE233 pKa = 3.7HH234 pKa = 7.02AIDD237 pKa = 4.0HH238 pKa = 5.69VQDD241 pKa = 4.66LLRR244 pKa = 11.84LPNKK248 pKa = 9.94LPRR251 pKa = 11.84VDD253 pKa = 5.57RR254 pKa = 11.84IPLEE258 pKa = 4.13GIRR261 pKa = 11.84YY262 pKa = 8.43DD263 pKa = 3.4KK264 pKa = 11.24SKK266 pKa = 11.07SCGAHH271 pKa = 5.28YY272 pKa = 10.41RR273 pKa = 11.84LQGFKK278 pKa = 10.11TRR280 pKa = 11.84GDD282 pKa = 3.26VWEE285 pKa = 4.11VAMFEE290 pKa = 4.39AKK292 pKa = 10.11NALVKK297 pKa = 10.65LWDD300 pKa = 3.81GEE302 pKa = 4.58YY303 pKa = 10.7VEE305 pKa = 5.04PRR307 pKa = 11.84PTRR310 pKa = 11.84MGGRR314 pKa = 11.84GKK316 pKa = 9.96LVKK319 pKa = 9.9MSQEE323 pKa = 3.68RR324 pKa = 11.84AVAEE328 pKa = 4.53GVAKK332 pKa = 10.47GRR334 pKa = 11.84AIHH337 pKa = 5.52MTDD340 pKa = 3.04TRR342 pKa = 11.84DD343 pKa = 3.67HH344 pKa = 6.97IILGLSEE351 pKa = 4.55QPLNDD356 pKa = 2.71AWKK359 pKa = 9.41PDD361 pKa = 3.64NFPISVGRR369 pKa = 11.84GWFHH373 pKa = 7.0GDD375 pKa = 2.53ATRR378 pKa = 11.84FVRR381 pKa = 11.84KK382 pKa = 8.79HH383 pKa = 5.75AGASRR388 pKa = 11.84IYY390 pKa = 10.84CFDD393 pKa = 3.39AEE395 pKa = 4.73KK396 pKa = 10.25FDD398 pKa = 4.21SSIMPWLIHH407 pKa = 5.77IAVTIMRR414 pKa = 11.84EE415 pKa = 3.69QFMEE419 pKa = 4.32GLRR422 pKa = 11.84HH423 pKa = 6.15DD424 pKa = 5.21ADD426 pKa = 4.11MYY428 pKa = 8.37WQFVEE433 pKa = 4.82EE434 pKa = 4.47SLLHH438 pKa = 7.06SMVFRR443 pKa = 11.84DD444 pKa = 4.73DD445 pKa = 3.68GVLFEE450 pKa = 5.35KK451 pKa = 11.12YY452 pKa = 9.68HH453 pKa = 5.45GTSSGHH459 pKa = 4.98NHH461 pKa = 6.0NSLAQSIVTCILASFNVFYY480 pKa = 10.88KK481 pKa = 10.69NRR483 pKa = 11.84EE484 pKa = 3.93LPVAVIKK491 pKa = 11.12ANFTIEE497 pKa = 4.3GLGDD501 pKa = 3.95DD502 pKa = 5.39NITCEE507 pKa = 4.17TDD509 pKa = 3.13VLADD513 pKa = 3.76EE514 pKa = 4.66TCEE517 pKa = 3.87EE518 pKa = 4.04RR519 pKa = 11.84GMRR522 pKa = 11.84TWNVFGVSWLGEE534 pKa = 3.77KK535 pKa = 10.34SFQTNTLCQPVVDD548 pKa = 3.81EE549 pKa = 5.23AEE551 pKa = 3.9WDD553 pKa = 3.7EE554 pKa = 4.26EE555 pKa = 4.68GMFGSAQYY563 pKa = 10.27LGKK566 pKa = 10.42FFRR569 pKa = 11.84EE570 pKa = 4.41DD571 pKa = 3.36VLEE574 pKa = 4.35VPSLGQVKK582 pKa = 9.16VVTPYY587 pKa = 10.04RR588 pKa = 11.84PQVEE592 pKa = 4.37TVLRR596 pKa = 11.84LLYY599 pKa = 10.16PEE601 pKa = 5.19GIMRR605 pKa = 11.84GDD607 pKa = 3.02AHH609 pKa = 7.63GDD611 pKa = 3.35FEE613 pKa = 5.22EE614 pKa = 4.48VFEE617 pKa = 4.25RR618 pKa = 11.84ARR620 pKa = 11.84GEE622 pKa = 3.99RR623 pKa = 11.84FAGHH627 pKa = 6.82ILDD630 pKa = 4.32GSGNPRR636 pKa = 11.84TRR638 pKa = 11.84AWLDD642 pKa = 3.9GYY644 pKa = 9.39WQFCHH649 pKa = 7.27DD650 pKa = 4.15NLLVIEE656 pKa = 4.39LTGHH660 pKa = 6.28HH661 pKa = 6.14SLEE664 pKa = 4.06KK665 pKa = 10.13RR666 pKa = 11.84LARR669 pKa = 11.84LGVEE673 pKa = 3.93IDD675 pKa = 3.42VDD677 pKa = 3.81DD678 pKa = 4.67AVYY681 pKa = 11.13DD682 pKa = 4.25DD683 pKa = 5.4GTAGWLRR690 pKa = 11.84LVNRR694 pKa = 11.84RR695 pKa = 11.84RR696 pKa = 11.84DD697 pKa = 3.87GNWEE701 pKa = 3.7FLQIVV706 pKa = 3.79
MM1 pKa = 7.67DD2 pKa = 4.7HH3 pKa = 7.13ARR5 pKa = 11.84DD6 pKa = 3.49FDD8 pKa = 4.73VISSNATNPLKK19 pKa = 10.58RR20 pKa = 11.84SRR22 pKa = 11.84EE23 pKa = 3.94EE24 pKa = 3.98ADD26 pKa = 3.39LGTAGEE32 pKa = 4.5APWRR36 pKa = 11.84TPVNGATINGVFRR49 pKa = 11.84IRR51 pKa = 11.84KK52 pKa = 9.16FDD54 pKa = 3.38VDD56 pKa = 4.0YY57 pKa = 11.63LNGPVADD64 pKa = 4.21AEE66 pKa = 4.58VVIAGLPRR74 pKa = 11.84VKK76 pKa = 10.31EE77 pKa = 4.15VVPEE81 pKa = 4.12SCDD84 pKa = 3.11ALRR87 pKa = 11.84GMKK90 pKa = 9.74VLNDD94 pKa = 3.64ADD96 pKa = 4.86HH97 pKa = 6.5ITTAVSSEE105 pKa = 4.22SLSGTGNGGTRR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84NSRR121 pKa = 11.84RR122 pKa = 11.84SRR124 pKa = 11.84KK125 pKa = 9.9NKK127 pKa = 9.7VKK129 pKa = 8.44TTAKK133 pKa = 10.37SGGSLDD139 pKa = 4.27DD140 pKa = 3.86LKK142 pKa = 11.68EE143 pKa = 4.07MFKK146 pKa = 10.76HH147 pKa = 5.1ITVHH151 pKa = 6.61DD152 pKa = 4.01AQVRR156 pKa = 11.84LGKK159 pKa = 10.24FHH161 pKa = 7.8DD162 pKa = 5.16EE163 pKa = 4.06ITFTPADD170 pKa = 3.95PAVLGYY176 pKa = 9.93LQSPEE181 pKa = 4.05GEE183 pKa = 4.01IPGFDD188 pKa = 3.33PSEE191 pKa = 4.22YY192 pKa = 10.69CFMEE196 pKa = 4.4ASGGVQMRR204 pKa = 11.84HH205 pKa = 5.06LKK207 pKa = 9.94MYY209 pKa = 10.92DD210 pKa = 3.35EE211 pKa = 4.93DD212 pKa = 4.22TSLGPDD218 pKa = 4.05FSLLEE223 pKa = 4.42DD224 pKa = 4.02PEE226 pKa = 4.75AGRR229 pKa = 11.84RR230 pKa = 11.84ALEE233 pKa = 3.7HH234 pKa = 7.02AIDD237 pKa = 4.0HH238 pKa = 5.69VQDD241 pKa = 4.66LLRR244 pKa = 11.84LPNKK248 pKa = 9.94LPRR251 pKa = 11.84VDD253 pKa = 5.57RR254 pKa = 11.84IPLEE258 pKa = 4.13GIRR261 pKa = 11.84YY262 pKa = 8.43DD263 pKa = 3.4KK264 pKa = 11.24SKK266 pKa = 11.07SCGAHH271 pKa = 5.28YY272 pKa = 10.41RR273 pKa = 11.84LQGFKK278 pKa = 10.11TRR280 pKa = 11.84GDD282 pKa = 3.26VWEE285 pKa = 4.11VAMFEE290 pKa = 4.39AKK292 pKa = 10.11NALVKK297 pKa = 10.65LWDD300 pKa = 3.81GEE302 pKa = 4.58YY303 pKa = 10.7VEE305 pKa = 5.04PRR307 pKa = 11.84PTRR310 pKa = 11.84MGGRR314 pKa = 11.84GKK316 pKa = 9.96LVKK319 pKa = 9.9MSQEE323 pKa = 3.68RR324 pKa = 11.84AVAEE328 pKa = 4.53GVAKK332 pKa = 10.47GRR334 pKa = 11.84AIHH337 pKa = 5.52MTDD340 pKa = 3.04TRR342 pKa = 11.84DD343 pKa = 3.67HH344 pKa = 6.97IILGLSEE351 pKa = 4.55QPLNDD356 pKa = 2.71AWKK359 pKa = 9.41PDD361 pKa = 3.64NFPISVGRR369 pKa = 11.84GWFHH373 pKa = 7.0GDD375 pKa = 2.53ATRR378 pKa = 11.84FVRR381 pKa = 11.84KK382 pKa = 8.79HH383 pKa = 5.75AGASRR388 pKa = 11.84IYY390 pKa = 10.84CFDD393 pKa = 3.39AEE395 pKa = 4.73KK396 pKa = 10.25FDD398 pKa = 4.21SSIMPWLIHH407 pKa = 5.77IAVTIMRR414 pKa = 11.84EE415 pKa = 3.69QFMEE419 pKa = 4.32GLRR422 pKa = 11.84HH423 pKa = 6.15DD424 pKa = 5.21ADD426 pKa = 4.11MYY428 pKa = 8.37WQFVEE433 pKa = 4.82EE434 pKa = 4.47SLLHH438 pKa = 7.06SMVFRR443 pKa = 11.84DD444 pKa = 4.73DD445 pKa = 3.68GVLFEE450 pKa = 5.35KK451 pKa = 11.12YY452 pKa = 9.68HH453 pKa = 5.45GTSSGHH459 pKa = 4.98NHH461 pKa = 6.0NSLAQSIVTCILASFNVFYY480 pKa = 10.88KK481 pKa = 10.69NRR483 pKa = 11.84EE484 pKa = 3.93LPVAVIKK491 pKa = 11.12ANFTIEE497 pKa = 4.3GLGDD501 pKa = 3.95DD502 pKa = 5.39NITCEE507 pKa = 4.17TDD509 pKa = 3.13VLADD513 pKa = 3.76EE514 pKa = 4.66TCEE517 pKa = 3.87EE518 pKa = 4.04RR519 pKa = 11.84GMRR522 pKa = 11.84TWNVFGVSWLGEE534 pKa = 3.77KK535 pKa = 10.34SFQTNTLCQPVVDD548 pKa = 3.81EE549 pKa = 5.23AEE551 pKa = 3.9WDD553 pKa = 3.7EE554 pKa = 4.26EE555 pKa = 4.68GMFGSAQYY563 pKa = 10.27LGKK566 pKa = 10.42FFRR569 pKa = 11.84EE570 pKa = 4.41DD571 pKa = 3.36VLEE574 pKa = 4.35VPSLGQVKK582 pKa = 9.16VVTPYY587 pKa = 10.04RR588 pKa = 11.84PQVEE592 pKa = 4.37TVLRR596 pKa = 11.84LLYY599 pKa = 10.16PEE601 pKa = 5.19GIMRR605 pKa = 11.84GDD607 pKa = 3.02AHH609 pKa = 7.63GDD611 pKa = 3.35FEE613 pKa = 5.22EE614 pKa = 4.48VFEE617 pKa = 4.25RR618 pKa = 11.84ARR620 pKa = 11.84GEE622 pKa = 3.99RR623 pKa = 11.84FAGHH627 pKa = 6.82ILDD630 pKa = 4.32GSGNPRR636 pKa = 11.84TRR638 pKa = 11.84AWLDD642 pKa = 3.9GYY644 pKa = 9.39WQFCHH649 pKa = 7.27DD650 pKa = 4.15NLLVIEE656 pKa = 4.39LTGHH660 pKa = 6.28HH661 pKa = 6.14SLEE664 pKa = 4.06KK665 pKa = 10.13RR666 pKa = 11.84LARR669 pKa = 11.84LGVEE673 pKa = 3.93IDD675 pKa = 3.42VDD677 pKa = 3.81DD678 pKa = 4.67AVYY681 pKa = 11.13DD682 pKa = 4.25DD683 pKa = 5.4GTAGWLRR690 pKa = 11.84LVNRR694 pKa = 11.84RR695 pKa = 11.84RR696 pKa = 11.84DD697 pKa = 3.87GNWEE701 pKa = 3.7FLQIVV706 pKa = 3.79
Molecular weight: 79.56 kDa
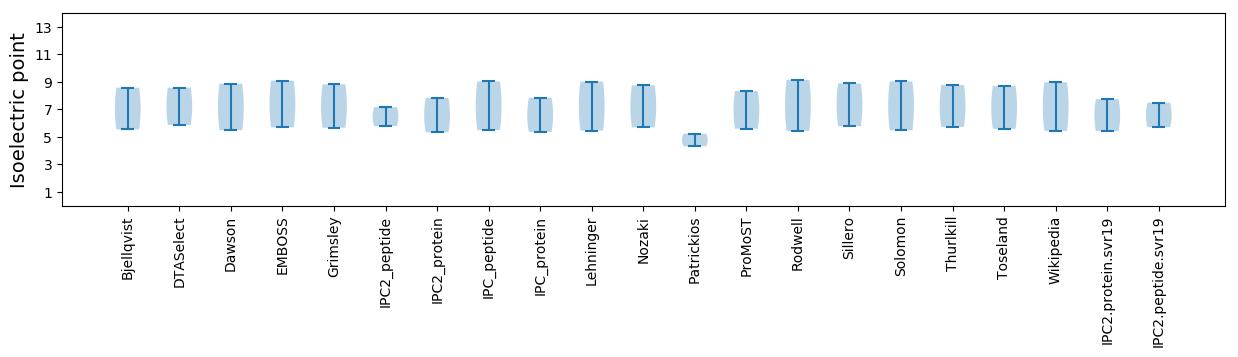
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F6Q4A5|A0A0F6Q4A5_9VIRU Putative RNA-dependent RNA polymerase OS=Rhizoctonia fumigata mycovirus OX=1592828 PE=4 SV=1
MM1 pKa = 7.27SGSMEE6 pKa = 4.39DD7 pKa = 4.46FEE9 pKa = 4.85ATLSRR14 pKa = 11.84VMGRR18 pKa = 11.84IGNVTNAPEE27 pKa = 3.76RR28 pKa = 11.84AEE30 pKa = 4.3TIEE33 pKa = 4.15EE34 pKa = 4.07AASQPRR40 pKa = 11.84APATTSVGVAKK51 pKa = 10.73EE52 pKa = 4.0EE53 pKa = 4.3DD54 pKa = 3.78SLPLVRR60 pKa = 11.84EE61 pKa = 3.97DD62 pKa = 3.38MTLKK66 pKa = 10.35EE67 pKa = 4.17LRR69 pKa = 11.84EE70 pKa = 3.96VMEE73 pKa = 4.52KK74 pKa = 10.13YY75 pKa = 10.68GKK77 pKa = 9.58VVEE80 pKa = 4.63RR81 pKa = 11.84GAYY84 pKa = 9.4LYY86 pKa = 10.28EE87 pKa = 3.96WTGGKK92 pKa = 9.13AAKK95 pKa = 10.48LNTLDD100 pKa = 3.74PKK102 pKa = 10.47EE103 pKa = 4.15LGRR106 pKa = 11.84EE107 pKa = 3.68RR108 pKa = 11.84MTSQEE113 pKa = 3.68RR114 pKa = 11.84RR115 pKa = 11.84ALDD118 pKa = 2.81WWEE121 pKa = 3.84EE122 pKa = 3.93KK123 pKa = 10.65GKK125 pKa = 10.88KK126 pKa = 9.72NEE128 pKa = 4.26FDD130 pKa = 3.12WTVCRR135 pKa = 11.84EE136 pKa = 4.1SGKK139 pKa = 10.28VPAGKK144 pKa = 9.3IGKK147 pKa = 9.71YY148 pKa = 9.43NDD150 pKa = 3.49EE151 pKa = 4.2LSAMRR156 pKa = 11.84WMYY159 pKa = 10.35QNKK162 pKa = 9.53ALTMEE167 pKa = 3.68NWFWLRR173 pKa = 11.84KK174 pKa = 7.74HH175 pKa = 6.25HH176 pKa = 6.07EE177 pKa = 4.3ARR179 pKa = 11.84SHH181 pKa = 5.26VVWALVKK188 pKa = 10.01IVKK191 pKa = 9.64AAQDD195 pKa = 3.6VTNGMDD201 pKa = 3.01TSDD204 pKa = 3.56ALGVAEE210 pKa = 4.41YY211 pKa = 8.74QTANKK216 pKa = 9.16IISIASTNVKK226 pKa = 9.03TFMEE230 pKa = 4.48QIHH233 pKa = 6.34RR234 pKa = 11.84LLRR237 pKa = 11.84PIDD240 pKa = 3.68EE241 pKa = 4.58GLNLVVARR249 pKa = 11.84KK250 pKa = 9.1IAMGEE255 pKa = 3.81RR256 pKa = 11.84LGIKK260 pKa = 8.39QKK262 pKa = 10.93KK263 pKa = 9.07VGTANRR269 pKa = 11.84QAAMFGEE276 pKa = 4.48EE277 pKa = 3.81AVGRR281 pKa = 11.84VTKK284 pKa = 10.47PSDD287 pKa = 3.36VPRR290 pKa = 11.84GTKK293 pKa = 8.45RR294 pKa = 11.84TRR296 pKa = 11.84DD297 pKa = 3.24EE298 pKa = 4.15TTTIDD303 pKa = 3.71ALRR306 pKa = 11.84KK307 pKa = 9.44LKK309 pKa = 10.65LSRR312 pKa = 11.84KK313 pKa = 8.04EE314 pKa = 4.03VVRR317 pKa = 11.84EE318 pKa = 3.83PAPEE322 pKa = 3.98NPPPVGRR329 pKa = 11.84PLVPQTMGDD338 pKa = 4.78DD339 pKa = 4.02GPEE342 pKa = 3.77WAPASPEE349 pKa = 4.03RR350 pKa = 11.84TDD352 pKa = 3.4VPLVKK357 pKa = 10.16SDD359 pKa = 3.66LTSAEE364 pKa = 3.92QKK366 pKa = 10.24IALDD370 pKa = 3.45WLKK373 pKa = 10.89KK374 pKa = 10.66GEE376 pKa = 4.26APTGAMGMAQEE387 pKa = 4.25WGVVAVRR394 pKa = 11.84SDD396 pKa = 3.43LLTRR400 pKa = 11.84AEE402 pKa = 4.23VKK404 pKa = 10.31KK405 pKa = 10.77GQGGKK410 pKa = 10.46LSLSVPKK417 pKa = 10.56GATRR421 pKa = 11.84AEE423 pKa = 4.37SVALASITMMMFRR436 pKa = 11.84GFLKK440 pKa = 10.37EE441 pKa = 4.75DD442 pKa = 3.49EE443 pKa = 4.4VPYY446 pKa = 10.22LLKK449 pKa = 10.33WADD452 pKa = 3.11LWGGLMRR459 pKa = 11.84YY460 pKa = 9.8AFGLTKK466 pKa = 10.46EE467 pKa = 4.31GVEE470 pKa = 4.24LLVQEE475 pKa = 4.47AMSAGEE481 pKa = 4.2VMGEE485 pKa = 3.86
MM1 pKa = 7.27SGSMEE6 pKa = 4.39DD7 pKa = 4.46FEE9 pKa = 4.85ATLSRR14 pKa = 11.84VMGRR18 pKa = 11.84IGNVTNAPEE27 pKa = 3.76RR28 pKa = 11.84AEE30 pKa = 4.3TIEE33 pKa = 4.15EE34 pKa = 4.07AASQPRR40 pKa = 11.84APATTSVGVAKK51 pKa = 10.73EE52 pKa = 4.0EE53 pKa = 4.3DD54 pKa = 3.78SLPLVRR60 pKa = 11.84EE61 pKa = 3.97DD62 pKa = 3.38MTLKK66 pKa = 10.35EE67 pKa = 4.17LRR69 pKa = 11.84EE70 pKa = 3.96VMEE73 pKa = 4.52KK74 pKa = 10.13YY75 pKa = 10.68GKK77 pKa = 9.58VVEE80 pKa = 4.63RR81 pKa = 11.84GAYY84 pKa = 9.4LYY86 pKa = 10.28EE87 pKa = 3.96WTGGKK92 pKa = 9.13AAKK95 pKa = 10.48LNTLDD100 pKa = 3.74PKK102 pKa = 10.47EE103 pKa = 4.15LGRR106 pKa = 11.84EE107 pKa = 3.68RR108 pKa = 11.84MTSQEE113 pKa = 3.68RR114 pKa = 11.84RR115 pKa = 11.84ALDD118 pKa = 2.81WWEE121 pKa = 3.84EE122 pKa = 3.93KK123 pKa = 10.65GKK125 pKa = 10.88KK126 pKa = 9.72NEE128 pKa = 4.26FDD130 pKa = 3.12WTVCRR135 pKa = 11.84EE136 pKa = 4.1SGKK139 pKa = 10.28VPAGKK144 pKa = 9.3IGKK147 pKa = 9.71YY148 pKa = 9.43NDD150 pKa = 3.49EE151 pKa = 4.2LSAMRR156 pKa = 11.84WMYY159 pKa = 10.35QNKK162 pKa = 9.53ALTMEE167 pKa = 3.68NWFWLRR173 pKa = 11.84KK174 pKa = 7.74HH175 pKa = 6.25HH176 pKa = 6.07EE177 pKa = 4.3ARR179 pKa = 11.84SHH181 pKa = 5.26VVWALVKK188 pKa = 10.01IVKK191 pKa = 9.64AAQDD195 pKa = 3.6VTNGMDD201 pKa = 3.01TSDD204 pKa = 3.56ALGVAEE210 pKa = 4.41YY211 pKa = 8.74QTANKK216 pKa = 9.16IISIASTNVKK226 pKa = 9.03TFMEE230 pKa = 4.48QIHH233 pKa = 6.34RR234 pKa = 11.84LLRR237 pKa = 11.84PIDD240 pKa = 3.68EE241 pKa = 4.58GLNLVVARR249 pKa = 11.84KK250 pKa = 9.1IAMGEE255 pKa = 3.81RR256 pKa = 11.84LGIKK260 pKa = 8.39QKK262 pKa = 10.93KK263 pKa = 9.07VGTANRR269 pKa = 11.84QAAMFGEE276 pKa = 4.48EE277 pKa = 3.81AVGRR281 pKa = 11.84VTKK284 pKa = 10.47PSDD287 pKa = 3.36VPRR290 pKa = 11.84GTKK293 pKa = 8.45RR294 pKa = 11.84TRR296 pKa = 11.84DD297 pKa = 3.24EE298 pKa = 4.15TTTIDD303 pKa = 3.71ALRR306 pKa = 11.84KK307 pKa = 9.44LKK309 pKa = 10.65LSRR312 pKa = 11.84KK313 pKa = 8.04EE314 pKa = 4.03VVRR317 pKa = 11.84EE318 pKa = 3.83PAPEE322 pKa = 3.98NPPPVGRR329 pKa = 11.84PLVPQTMGDD338 pKa = 4.78DD339 pKa = 4.02GPEE342 pKa = 3.77WAPASPEE349 pKa = 4.03RR350 pKa = 11.84TDD352 pKa = 3.4VPLVKK357 pKa = 10.16SDD359 pKa = 3.66LTSAEE364 pKa = 3.92QKK366 pKa = 10.24IALDD370 pKa = 3.45WLKK373 pKa = 10.89KK374 pKa = 10.66GEE376 pKa = 4.26APTGAMGMAQEE387 pKa = 4.25WGVVAVRR394 pKa = 11.84SDD396 pKa = 3.43LLTRR400 pKa = 11.84AEE402 pKa = 4.23VKK404 pKa = 10.31KK405 pKa = 10.77GQGGKK410 pKa = 10.46LSLSVPKK417 pKa = 10.56GATRR421 pKa = 11.84AEE423 pKa = 4.37SVALASITMMMFRR436 pKa = 11.84GFLKK440 pKa = 10.37EE441 pKa = 4.75DD442 pKa = 3.49EE443 pKa = 4.4VPYY446 pKa = 10.22LLKK449 pKa = 10.33WADD452 pKa = 3.11LWGGLMRR459 pKa = 11.84YY460 pKa = 9.8AFGLTKK466 pKa = 10.46EE467 pKa = 4.31GVEE470 pKa = 4.24LLVQEE475 pKa = 4.47AMSAGEE481 pKa = 4.2VMGEE485 pKa = 3.86
Molecular weight: 53.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1191 |
485 |
706 |
595.5 |
66.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.144 ± 1.045 | 0.84 ± 0.378 |
6.465 ± 1.15 | 8.564 ± 0.795 |
3.442 ± 1.069 | 8.564 ± 0.189 |
2.351 ± 0.91 | 3.694 ± 0.482 |
5.877 ± 1.29 | 8.312 ± 0.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.359 ± 0.702 | 3.191 ± 0.304 |
4.366 ± 0.224 | 2.519 ± 0.096 |
7.473 ± 0.276 | 5.206 ± 0.153 |
5.458 ± 0.557 | 7.893 ± 0.089 |
2.267 ± 0.246 | 2.015 ± 0.218 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
