
Fimbriiglobus ruber
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Gemmatales; Gemmataceae; Fimbriiglobus
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
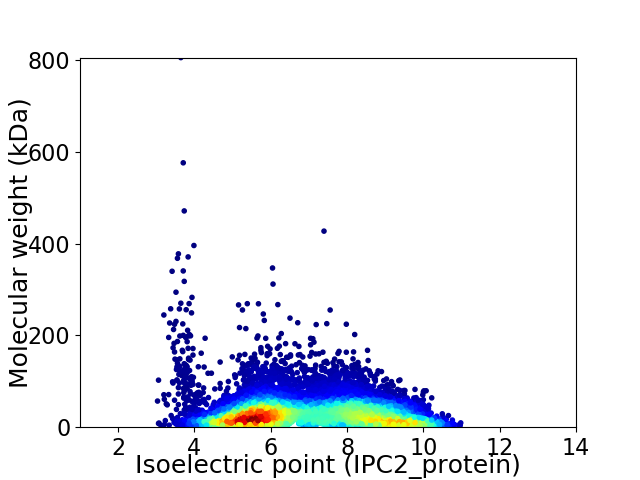
Virtual 2D-PAGE plot for 10212 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A225E089|A0A225E089_9BACT Peptidase_M78 domain-containing protein OS=Fimbriiglobus ruber OX=1908690 GN=FRUB_00661 PE=4 SV=1
MM1 pKa = 7.54IRR3 pKa = 11.84NDD5 pKa = 3.57WRR7 pKa = 11.84RR8 pKa = 11.84MIQRR12 pKa = 11.84VGRR15 pKa = 11.84GQHH18 pKa = 5.11KK19 pKa = 9.16QNRR22 pKa = 11.84SPSMTVLQLEE32 pKa = 4.55DD33 pKa = 3.73RR34 pKa = 11.84SVPATFTAGSIQGQDD49 pKa = 2.62GWTGGTAAISTNVVQTVDD67 pKa = 3.3QSGANAFSGTGAWLVSNSTANGGYY91 pKa = 9.92NGNFVGWPFSPGLSVTAGDD110 pKa = 4.01PSSGAGADD118 pKa = 3.43TFTATLYY125 pKa = 10.54FKK127 pKa = 10.47SASSAADD134 pKa = 3.46GSNIEE139 pKa = 4.09VDD141 pKa = 3.75LGSAAGDD148 pKa = 3.45DD149 pKa = 3.66RR150 pKa = 11.84TNFLALTNEE159 pKa = 4.1DD160 pKa = 4.23DD161 pKa = 4.84ANGGLQIRR169 pKa = 11.84AAEE172 pKa = 4.12PMTDD176 pKa = 3.04GTTNFYY182 pKa = 8.51PTVSIATDD190 pKa = 3.18ITRR193 pKa = 11.84ATYY196 pKa = 10.22HH197 pKa = 6.61RR198 pKa = 11.84LDD200 pKa = 3.29VTANYY205 pKa = 11.0ADD207 pKa = 3.86GTDD210 pKa = 3.27NDD212 pKa = 4.53TFTVSLDD219 pKa = 3.45GKK221 pKa = 10.48VLTNPRR227 pKa = 11.84TGGTTFGTFEE237 pKa = 4.49AYY239 pKa = 8.47EE240 pKa = 4.22AAASPPASPPNYY252 pKa = 8.69EE253 pKa = 3.74QTSRR257 pKa = 11.84LLFRR261 pKa = 11.84SGTAPSGFDD270 pKa = 3.22ALTSPGEE277 pKa = 4.08FSDD280 pKa = 3.66TGAQGFYY287 pKa = 10.19FDD289 pKa = 3.62NVSYY293 pKa = 10.97RR294 pKa = 11.84DD295 pKa = 3.89YY296 pKa = 11.12NQSDD300 pKa = 3.76PSATLASYY308 pKa = 10.55AATFEE313 pKa = 4.25TQPAVTYY320 pKa = 10.52ANPAWSSLATGDD332 pKa = 5.02PITDD336 pKa = 4.28ADD338 pKa = 4.47PLTAGDD344 pKa = 3.76QPATYY349 pKa = 9.84GVNAFATITDD359 pKa = 3.53AVTAVAAGGTVIANAGTYY377 pKa = 10.26AEE379 pKa = 4.44NVTINKK385 pKa = 9.24PLTLEE390 pKa = 4.52GQSSTNTVIEE400 pKa = 4.46PGLTSSYY407 pKa = 9.75NTDD410 pKa = 2.93NVVTVLANNVTVEE423 pKa = 3.99GFTIQGSITGTPPVGQSSGFTLSSGTTVYY452 pKa = 10.84AAAGISNSADD462 pKa = 3.36VTSTATDD469 pKa = 3.1ISGLTVQNNVIKK481 pKa = 10.73DD482 pKa = 3.56FTQFGVDD489 pKa = 3.46GDD491 pKa = 4.2TSSGAVSTGNTIANNVIQDD510 pKa = 3.52IPEE513 pKa = 4.31NNPGSGFYY521 pKa = 10.62GQGVVVADD529 pKa = 3.6NFYY532 pKa = 11.3AAITGNTITNVRR544 pKa = 11.84TGIQTDD550 pKa = 3.37NNYY553 pKa = 9.63TSAGAFAPSISNNIVSATVKK573 pKa = 10.64GIYY576 pKa = 10.46LNLQYY581 pKa = 11.07SAASPFAITGNTITQADD598 pKa = 4.31GTVSPAYY605 pKa = 10.28NVGLLVQSIYY615 pKa = 11.42NGVTTNIQGNNVSGFLYY632 pKa = 10.2GVEE635 pKa = 4.11FAGNSATTPVEE646 pKa = 4.19VQGGTLSGNTYY657 pKa = 10.28GVWATNNDD665 pKa = 3.2YY666 pKa = 10.72FYY668 pKa = 10.24PANFNTTAALSGVAITDD685 pKa = 3.67SKK687 pKa = 9.86TAAIWVDD694 pKa = 3.41STSQSMAGTSDD705 pKa = 3.2TTDD708 pKa = 3.24TVSLAVGGGTTVTGGPTGLLIAGSLSNITGNALGTVSFAGVSGDD752 pKa = 5.36DD753 pKa = 3.12ITLAAGALVGQTLDD767 pKa = 3.57ATAATYY773 pKa = 10.82DD774 pKa = 3.97GVVGTAATPTQGYY787 pKa = 10.22AIADD791 pKa = 3.85KK792 pKa = 10.37VTDD795 pKa = 4.34YY796 pKa = 11.23IDD798 pKa = 3.73NPALGFVRR806 pKa = 11.84ITPATVYY813 pKa = 7.48VTPKK817 pKa = 10.35SEE819 pKa = 4.11STTAGAVQRR828 pKa = 11.84GVTAAANNDD837 pKa = 3.89TVDD840 pKa = 3.63VAAGTYY846 pKa = 10.33AEE848 pKa = 4.49NVTINKK854 pKa = 9.25PLTLEE859 pKa = 4.6GEE861 pKa = 4.41SSANTVIEE869 pKa = 4.41PGLTSSYY876 pKa = 9.75NTDD879 pKa = 2.93NVVTVLANDD888 pKa = 3.39VTVEE892 pKa = 3.95GFTIQGSITGTPPVGQSSGFTLSSGTTVYY921 pKa = 10.84AAAGISNSADD931 pKa = 3.36VTSTATDD938 pKa = 3.1ISGLTVQNNVIKK950 pKa = 10.73DD951 pKa = 3.56FTQFGVDD958 pKa = 3.46GDD960 pKa = 4.2TSSGAVSTGNTIANNVIQDD979 pKa = 3.52IPEE982 pKa = 4.31NNPGSGFYY990 pKa = 10.62GQGVVVADD998 pKa = 3.6NFYY1001 pKa = 11.3AAITGNTITNVRR1013 pKa = 11.84TGIQTDD1019 pKa = 3.37NNYY1022 pKa = 9.63TSAGAFAPSISNNIVSATVKK1042 pKa = 10.64GIYY1045 pKa = 10.46LNLQYY1050 pKa = 11.07SAASPFAITGNTITQADD1067 pKa = 4.31GTVSPAYY1074 pKa = 10.28NVGLLVQSIYY1084 pKa = 11.42NGVTTNIQGNNVSGFLYY1101 pKa = 10.2GVEE1104 pKa = 4.11FAGNSATTPVEE1115 pKa = 4.19VQGGTLSGNTYY1126 pKa = 10.28GVWATNNDD1134 pKa = 3.2YY1135 pKa = 10.72FYY1137 pKa = 10.24PANFNTTAALSGVAITDD1154 pKa = 3.67SKK1156 pKa = 9.86TAAIWVDD1163 pKa = 3.41STSQSMAGTSDD1174 pKa = 3.25TTDD1177 pKa = 3.14SVSLAVGGGTSVTGGPTGLLIAGGLSNITGNALGTVSFAGVTGDD1221 pKa = 5.59DD1222 pKa = 3.15ITLAAGALAGQTLDD1236 pKa = 3.42ATGVTFNGTTGSTATPAQDD1255 pKa = 3.29YY1256 pKa = 11.03AIADD1260 pKa = 4.37KK1261 pKa = 10.14ITDD1264 pKa = 4.81FIDD1267 pKa = 4.46DD1268 pKa = 3.79PTLGFVQITPATVYY1282 pKa = 7.54VTPKK1286 pKa = 10.46SEE1288 pKa = 3.9ATTAGAIPRR1297 pKa = 11.84AVTVASAGDD1306 pKa = 4.1TIDD1309 pKa = 3.61VAAGTYY1315 pKa = 8.49TGNIDD1320 pKa = 3.3LSKK1323 pKa = 10.45NVSVRR1328 pKa = 11.84IDD1330 pKa = 3.71GPSSLTGAISGTTGSITDD1348 pKa = 3.52TLGAVSLGDD1357 pKa = 3.57GSAAGVATSSTLDD1370 pKa = 3.37VNGQTVTLLDD1380 pKa = 3.75SDD1382 pKa = 4.58GADD1385 pKa = 3.69LDD1387 pKa = 4.53GEE1389 pKa = 4.41TDD1391 pKa = 3.66LVGGTLVSANGINVGGTLAGPGTVGAATIQAGGNVEE1427 pKa = 4.24PGGSGTGILNTGNIAFAAGATLTPTLKK1454 pKa = 9.9GTTAGTTYY1462 pKa = 10.91DD1463 pKa = 3.47QVNTTGTVDD1472 pKa = 4.3LGGATLNLNLAFTPAAGDD1490 pKa = 3.28TFTIVNNDD1498 pKa = 2.7GTDD1501 pKa = 3.64PVVGTFAGLPEE1512 pKa = 4.24GSVFKK1517 pKa = 11.01VGSQFFQISYY1527 pKa = 10.53KK1528 pKa = 10.89GGDD1531 pKa = 3.81GNDD1534 pKa = 3.38VTLKK1538 pKa = 10.55AVPSSVLISAPSAAFANDD1556 pKa = 3.26TSTITYY1562 pKa = 7.21TVTYY1566 pKa = 9.73ADD1568 pKa = 4.3PNFASSTLSSGDD1580 pKa = 3.15VTLNTTGTAAVGSVVVSGTGATRR1603 pKa = 11.84TVTLSNITGDD1613 pKa = 3.74GAVGISLPAGTGTDD1627 pKa = 3.22TSSNPEE1633 pKa = 4.01PGAGPSATFTADD1645 pKa = 2.85STAPTVAIGAPSAAFANGTSTVTYY1669 pKa = 9.06TITYY1673 pKa = 8.23TDD1675 pKa = 3.45TNFADD1680 pKa = 3.84STLTPGDD1687 pKa = 3.39VTLNATGDD1695 pKa = 3.65ANATISIDD1703 pKa = 3.43SGTGATRR1710 pKa = 11.84TVTLTHH1716 pKa = 5.09VTGTGTLGISLLADD1730 pKa = 3.48TAVDD1734 pKa = 3.55SAGNEE1739 pKa = 3.75APAAGPSGTFTADD1752 pKa = 2.97TTNPTVVIGAPSAATANAGSTITYY1776 pKa = 7.58TITYY1780 pKa = 9.67SDD1782 pKa = 3.81TNFDD1786 pKa = 3.74QSTLTAADD1794 pKa = 3.39VTLNTTGTASATVTVDD1810 pKa = 3.2GGAGATRR1817 pKa = 11.84TVTLTHH1823 pKa = 5.96ITGDD1827 pKa = 3.47GTLGISLAAGTAHH1840 pKa = 7.56DD1841 pKa = 4.43LAGNEE1846 pKa = 4.27APVAGPSGTFTVDD1859 pKa = 2.87TTAPTVAIGAPSAALANAASTVTFTVTYY1887 pKa = 10.89ADD1889 pKa = 3.74ANFAASTLTAADD1901 pKa = 3.43VTLNTTGTAAGTVSVSGSGTTWTVTVTGITGDD1933 pKa = 3.53GTLGISLAAGTASDD1947 pKa = 3.8TAGNLAPAAGPSATFTVDD1965 pKa = 2.97DD1966 pKa = 4.05TAPTVSISPPSVTVANGSSTVTYY1989 pKa = 9.33TVTYY1993 pKa = 10.78ADD1995 pKa = 4.39ANFADD2000 pKa = 4.28STLTAADD2007 pKa = 3.44VTLNATGTASATVSVDD2023 pKa = 3.01SGSGTTRR2030 pKa = 11.84TVTLTHH2036 pKa = 5.96ITGDD2040 pKa = 3.47GTLGISLAAGTAHH2053 pKa = 7.11DD2054 pKa = 4.52QLSDD2058 pKa = 3.73PAPAAGPSGTFAVDD2072 pKa = 3.45TTAPTVAIGNPSVAITNGSSTVTYY2096 pKa = 9.6TITYY2100 pKa = 10.04SDD2102 pKa = 4.03ANFAASTLTAADD2114 pKa = 3.43VTLNTTGTATATVSVDD2130 pKa = 2.9SGTGTTRR2137 pKa = 11.84TITLSNITGDD2147 pKa = 3.48GTLGISLVAGTASDD2161 pKa = 3.48AAEE2164 pKa = 4.33NLAPAAGPSGTFAVDD2179 pKa = 3.45TTAPTVTISSPAVTGAGAAEE2199 pKa = 4.23VVTYY2203 pKa = 9.3TITYY2207 pKa = 10.34ADD2209 pKa = 4.01ANFADD2214 pKa = 4.28STLTAADD2221 pKa = 3.44VTLNATGGTTASVAVSGTGTTRR2243 pKa = 11.84TVTLTHH2249 pKa = 6.03ITGSGTVGISLVSGTAVDD2267 pKa = 3.61TAGNLAPAAGPSATAAAVPVDD2288 pKa = 4.07ANAVGQFAASPDD2300 pKa = 3.59AGGSGIVTVYY2310 pKa = 10.75NADD2313 pKa = 3.16GSVAYY2318 pKa = 8.44TVNPFPGVGAQTGVRR2333 pKa = 11.84AVMAGVTGSATPDD2346 pKa = 3.43VIAGTGPGVRR2356 pKa = 11.84AQVVVVSGTTHH2367 pKa = 5.92AVVMTLNPFEE2377 pKa = 5.56DD2378 pKa = 4.21SFTGGVFVAAADD2390 pKa = 4.12LNGDD2394 pKa = 3.6GHH2396 pKa = 8.59ADD2398 pKa = 2.99IVVSPDD2404 pKa = 2.88VGGGGRR2410 pKa = 11.84ITVYY2414 pKa = 10.72DD2415 pKa = 3.89GATGQVIANFFGIDD2429 pKa = 3.37DD2430 pKa = 3.83PAFRR2434 pKa = 11.84GGARR2438 pKa = 11.84VSFGDD2443 pKa = 3.56VNGDD2447 pKa = 3.63GVPDD2451 pKa = 4.93LIVSAGTGGGPRR2463 pKa = 11.84VAIFDD2468 pKa = 3.69GRR2470 pKa = 11.84SIGLGKK2476 pKa = 8.74TPTKK2480 pKa = 10.53LVADD2484 pKa = 5.11FFAFEE2489 pKa = 3.85PSLRR2493 pKa = 11.84NGAYY2497 pKa = 9.55VAAGDD2502 pKa = 4.18FNGDD2506 pKa = 3.09GYY2508 pKa = 11.81ADD2510 pKa = 4.2LVAGGGPDD2518 pKa = 3.31GAPRR2522 pKa = 11.84VEE2524 pKa = 4.3VLDD2527 pKa = 4.25GASLLSSNGQAPTMVANFFAGDD2549 pKa = 3.42PSLRR2553 pKa = 11.84SGARR2557 pKa = 11.84VAVKK2561 pKa = 10.56NLDD2564 pKa = 3.58GDD2566 pKa = 4.11GSADD2570 pKa = 3.72LVVGLATGTGTQVATYY2586 pKa = 10.0LGKK2589 pKa = 10.33DD2590 pKa = 3.41LTASGTPTAAAEE2602 pKa = 4.1LDD2604 pKa = 3.93PFPGFTGGVFVGG2616 pKa = 4.17
MM1 pKa = 7.54IRR3 pKa = 11.84NDD5 pKa = 3.57WRR7 pKa = 11.84RR8 pKa = 11.84MIQRR12 pKa = 11.84VGRR15 pKa = 11.84GQHH18 pKa = 5.11KK19 pKa = 9.16QNRR22 pKa = 11.84SPSMTVLQLEE32 pKa = 4.55DD33 pKa = 3.73RR34 pKa = 11.84SVPATFTAGSIQGQDD49 pKa = 2.62GWTGGTAAISTNVVQTVDD67 pKa = 3.3QSGANAFSGTGAWLVSNSTANGGYY91 pKa = 9.92NGNFVGWPFSPGLSVTAGDD110 pKa = 4.01PSSGAGADD118 pKa = 3.43TFTATLYY125 pKa = 10.54FKK127 pKa = 10.47SASSAADD134 pKa = 3.46GSNIEE139 pKa = 4.09VDD141 pKa = 3.75LGSAAGDD148 pKa = 3.45DD149 pKa = 3.66RR150 pKa = 11.84TNFLALTNEE159 pKa = 4.1DD160 pKa = 4.23DD161 pKa = 4.84ANGGLQIRR169 pKa = 11.84AAEE172 pKa = 4.12PMTDD176 pKa = 3.04GTTNFYY182 pKa = 8.51PTVSIATDD190 pKa = 3.18ITRR193 pKa = 11.84ATYY196 pKa = 10.22HH197 pKa = 6.61RR198 pKa = 11.84LDD200 pKa = 3.29VTANYY205 pKa = 11.0ADD207 pKa = 3.86GTDD210 pKa = 3.27NDD212 pKa = 4.53TFTVSLDD219 pKa = 3.45GKK221 pKa = 10.48VLTNPRR227 pKa = 11.84TGGTTFGTFEE237 pKa = 4.49AYY239 pKa = 8.47EE240 pKa = 4.22AAASPPASPPNYY252 pKa = 8.69EE253 pKa = 3.74QTSRR257 pKa = 11.84LLFRR261 pKa = 11.84SGTAPSGFDD270 pKa = 3.22ALTSPGEE277 pKa = 4.08FSDD280 pKa = 3.66TGAQGFYY287 pKa = 10.19FDD289 pKa = 3.62NVSYY293 pKa = 10.97RR294 pKa = 11.84DD295 pKa = 3.89YY296 pKa = 11.12NQSDD300 pKa = 3.76PSATLASYY308 pKa = 10.55AATFEE313 pKa = 4.25TQPAVTYY320 pKa = 10.52ANPAWSSLATGDD332 pKa = 5.02PITDD336 pKa = 4.28ADD338 pKa = 4.47PLTAGDD344 pKa = 3.76QPATYY349 pKa = 9.84GVNAFATITDD359 pKa = 3.53AVTAVAAGGTVIANAGTYY377 pKa = 10.26AEE379 pKa = 4.44NVTINKK385 pKa = 9.24PLTLEE390 pKa = 4.52GQSSTNTVIEE400 pKa = 4.46PGLTSSYY407 pKa = 9.75NTDD410 pKa = 2.93NVVTVLANNVTVEE423 pKa = 3.99GFTIQGSITGTPPVGQSSGFTLSSGTTVYY452 pKa = 10.84AAAGISNSADD462 pKa = 3.36VTSTATDD469 pKa = 3.1ISGLTVQNNVIKK481 pKa = 10.73DD482 pKa = 3.56FTQFGVDD489 pKa = 3.46GDD491 pKa = 4.2TSSGAVSTGNTIANNVIQDD510 pKa = 3.52IPEE513 pKa = 4.31NNPGSGFYY521 pKa = 10.62GQGVVVADD529 pKa = 3.6NFYY532 pKa = 11.3AAITGNTITNVRR544 pKa = 11.84TGIQTDD550 pKa = 3.37NNYY553 pKa = 9.63TSAGAFAPSISNNIVSATVKK573 pKa = 10.64GIYY576 pKa = 10.46LNLQYY581 pKa = 11.07SAASPFAITGNTITQADD598 pKa = 4.31GTVSPAYY605 pKa = 10.28NVGLLVQSIYY615 pKa = 11.42NGVTTNIQGNNVSGFLYY632 pKa = 10.2GVEE635 pKa = 4.11FAGNSATTPVEE646 pKa = 4.19VQGGTLSGNTYY657 pKa = 10.28GVWATNNDD665 pKa = 3.2YY666 pKa = 10.72FYY668 pKa = 10.24PANFNTTAALSGVAITDD685 pKa = 3.67SKK687 pKa = 9.86TAAIWVDD694 pKa = 3.41STSQSMAGTSDD705 pKa = 3.2TTDD708 pKa = 3.24TVSLAVGGGTTVTGGPTGLLIAGSLSNITGNALGTVSFAGVSGDD752 pKa = 5.36DD753 pKa = 3.12ITLAAGALVGQTLDD767 pKa = 3.57ATAATYY773 pKa = 10.82DD774 pKa = 3.97GVVGTAATPTQGYY787 pKa = 10.22AIADD791 pKa = 3.85KK792 pKa = 10.37VTDD795 pKa = 4.34YY796 pKa = 11.23IDD798 pKa = 3.73NPALGFVRR806 pKa = 11.84ITPATVYY813 pKa = 7.48VTPKK817 pKa = 10.35SEE819 pKa = 4.11STTAGAVQRR828 pKa = 11.84GVTAAANNDD837 pKa = 3.89TVDD840 pKa = 3.63VAAGTYY846 pKa = 10.33AEE848 pKa = 4.49NVTINKK854 pKa = 9.25PLTLEE859 pKa = 4.6GEE861 pKa = 4.41SSANTVIEE869 pKa = 4.41PGLTSSYY876 pKa = 9.75NTDD879 pKa = 2.93NVVTVLANDD888 pKa = 3.39VTVEE892 pKa = 3.95GFTIQGSITGTPPVGQSSGFTLSSGTTVYY921 pKa = 10.84AAAGISNSADD931 pKa = 3.36VTSTATDD938 pKa = 3.1ISGLTVQNNVIKK950 pKa = 10.73DD951 pKa = 3.56FTQFGVDD958 pKa = 3.46GDD960 pKa = 4.2TSSGAVSTGNTIANNVIQDD979 pKa = 3.52IPEE982 pKa = 4.31NNPGSGFYY990 pKa = 10.62GQGVVVADD998 pKa = 3.6NFYY1001 pKa = 11.3AAITGNTITNVRR1013 pKa = 11.84TGIQTDD1019 pKa = 3.37NNYY1022 pKa = 9.63TSAGAFAPSISNNIVSATVKK1042 pKa = 10.64GIYY1045 pKa = 10.46LNLQYY1050 pKa = 11.07SAASPFAITGNTITQADD1067 pKa = 4.31GTVSPAYY1074 pKa = 10.28NVGLLVQSIYY1084 pKa = 11.42NGVTTNIQGNNVSGFLYY1101 pKa = 10.2GVEE1104 pKa = 4.11FAGNSATTPVEE1115 pKa = 4.19VQGGTLSGNTYY1126 pKa = 10.28GVWATNNDD1134 pKa = 3.2YY1135 pKa = 10.72FYY1137 pKa = 10.24PANFNTTAALSGVAITDD1154 pKa = 3.67SKK1156 pKa = 9.86TAAIWVDD1163 pKa = 3.41STSQSMAGTSDD1174 pKa = 3.25TTDD1177 pKa = 3.14SVSLAVGGGTSVTGGPTGLLIAGGLSNITGNALGTVSFAGVTGDD1221 pKa = 5.59DD1222 pKa = 3.15ITLAAGALAGQTLDD1236 pKa = 3.42ATGVTFNGTTGSTATPAQDD1255 pKa = 3.29YY1256 pKa = 11.03AIADD1260 pKa = 4.37KK1261 pKa = 10.14ITDD1264 pKa = 4.81FIDD1267 pKa = 4.46DD1268 pKa = 3.79PTLGFVQITPATVYY1282 pKa = 7.54VTPKK1286 pKa = 10.46SEE1288 pKa = 3.9ATTAGAIPRR1297 pKa = 11.84AVTVASAGDD1306 pKa = 4.1TIDD1309 pKa = 3.61VAAGTYY1315 pKa = 8.49TGNIDD1320 pKa = 3.3LSKK1323 pKa = 10.45NVSVRR1328 pKa = 11.84IDD1330 pKa = 3.71GPSSLTGAISGTTGSITDD1348 pKa = 3.52TLGAVSLGDD1357 pKa = 3.57GSAAGVATSSTLDD1370 pKa = 3.37VNGQTVTLLDD1380 pKa = 3.75SDD1382 pKa = 4.58GADD1385 pKa = 3.69LDD1387 pKa = 4.53GEE1389 pKa = 4.41TDD1391 pKa = 3.66LVGGTLVSANGINVGGTLAGPGTVGAATIQAGGNVEE1427 pKa = 4.24PGGSGTGILNTGNIAFAAGATLTPTLKK1454 pKa = 9.9GTTAGTTYY1462 pKa = 10.91DD1463 pKa = 3.47QVNTTGTVDD1472 pKa = 4.3LGGATLNLNLAFTPAAGDD1490 pKa = 3.28TFTIVNNDD1498 pKa = 2.7GTDD1501 pKa = 3.64PVVGTFAGLPEE1512 pKa = 4.24GSVFKK1517 pKa = 11.01VGSQFFQISYY1527 pKa = 10.53KK1528 pKa = 10.89GGDD1531 pKa = 3.81GNDD1534 pKa = 3.38VTLKK1538 pKa = 10.55AVPSSVLISAPSAAFANDD1556 pKa = 3.26TSTITYY1562 pKa = 7.21TVTYY1566 pKa = 9.73ADD1568 pKa = 4.3PNFASSTLSSGDD1580 pKa = 3.15VTLNTTGTAAVGSVVVSGTGATRR1603 pKa = 11.84TVTLSNITGDD1613 pKa = 3.74GAVGISLPAGTGTDD1627 pKa = 3.22TSSNPEE1633 pKa = 4.01PGAGPSATFTADD1645 pKa = 2.85STAPTVAIGAPSAAFANGTSTVTYY1669 pKa = 9.06TITYY1673 pKa = 8.23TDD1675 pKa = 3.45TNFADD1680 pKa = 3.84STLTPGDD1687 pKa = 3.39VTLNATGDD1695 pKa = 3.65ANATISIDD1703 pKa = 3.43SGTGATRR1710 pKa = 11.84TVTLTHH1716 pKa = 5.09VTGTGTLGISLLADD1730 pKa = 3.48TAVDD1734 pKa = 3.55SAGNEE1739 pKa = 3.75APAAGPSGTFTADD1752 pKa = 2.97TTNPTVVIGAPSAATANAGSTITYY1776 pKa = 7.58TITYY1780 pKa = 9.67SDD1782 pKa = 3.81TNFDD1786 pKa = 3.74QSTLTAADD1794 pKa = 3.39VTLNTTGTASATVTVDD1810 pKa = 3.2GGAGATRR1817 pKa = 11.84TVTLTHH1823 pKa = 5.96ITGDD1827 pKa = 3.47GTLGISLAAGTAHH1840 pKa = 7.56DD1841 pKa = 4.43LAGNEE1846 pKa = 4.27APVAGPSGTFTVDD1859 pKa = 2.87TTAPTVAIGAPSAALANAASTVTFTVTYY1887 pKa = 10.89ADD1889 pKa = 3.74ANFAASTLTAADD1901 pKa = 3.43VTLNTTGTAAGTVSVSGSGTTWTVTVTGITGDD1933 pKa = 3.53GTLGISLAAGTASDD1947 pKa = 3.8TAGNLAPAAGPSATFTVDD1965 pKa = 2.97DD1966 pKa = 4.05TAPTVSISPPSVTVANGSSTVTYY1989 pKa = 9.33TVTYY1993 pKa = 10.78ADD1995 pKa = 4.39ANFADD2000 pKa = 4.28STLTAADD2007 pKa = 3.44VTLNATGTASATVSVDD2023 pKa = 3.01SGSGTTRR2030 pKa = 11.84TVTLTHH2036 pKa = 5.96ITGDD2040 pKa = 3.47GTLGISLAAGTAHH2053 pKa = 7.11DD2054 pKa = 4.52QLSDD2058 pKa = 3.73PAPAAGPSGTFAVDD2072 pKa = 3.45TTAPTVAIGNPSVAITNGSSTVTYY2096 pKa = 9.6TITYY2100 pKa = 10.04SDD2102 pKa = 4.03ANFAASTLTAADD2114 pKa = 3.43VTLNTTGTATATVSVDD2130 pKa = 2.9SGTGTTRR2137 pKa = 11.84TITLSNITGDD2147 pKa = 3.48GTLGISLVAGTASDD2161 pKa = 3.48AAEE2164 pKa = 4.33NLAPAAGPSGTFAVDD2179 pKa = 3.45TTAPTVTISSPAVTGAGAAEE2199 pKa = 4.23VVTYY2203 pKa = 9.3TITYY2207 pKa = 10.34ADD2209 pKa = 4.01ANFADD2214 pKa = 4.28STLTAADD2221 pKa = 3.44VTLNATGGTTASVAVSGTGTTRR2243 pKa = 11.84TVTLTHH2249 pKa = 6.03ITGSGTVGISLVSGTAVDD2267 pKa = 3.61TAGNLAPAAGPSATAAAVPVDD2288 pKa = 4.07ANAVGQFAASPDD2300 pKa = 3.59AGGSGIVTVYY2310 pKa = 10.75NADD2313 pKa = 3.16GSVAYY2318 pKa = 8.44TVNPFPGVGAQTGVRR2333 pKa = 11.84AVMAGVTGSATPDD2346 pKa = 3.43VIAGTGPGVRR2356 pKa = 11.84AQVVVVSGTTHH2367 pKa = 5.92AVVMTLNPFEE2377 pKa = 5.56DD2378 pKa = 4.21SFTGGVFVAAADD2390 pKa = 4.12LNGDD2394 pKa = 3.6GHH2396 pKa = 8.59ADD2398 pKa = 2.99IVVSPDD2404 pKa = 2.88VGGGGRR2410 pKa = 11.84ITVYY2414 pKa = 10.72DD2415 pKa = 3.89GATGQVIANFFGIDD2429 pKa = 3.37DD2430 pKa = 3.83PAFRR2434 pKa = 11.84GGARR2438 pKa = 11.84VSFGDD2443 pKa = 3.56VNGDD2447 pKa = 3.63GVPDD2451 pKa = 4.93LIVSAGTGGGPRR2463 pKa = 11.84VAIFDD2468 pKa = 3.69GRR2470 pKa = 11.84SIGLGKK2476 pKa = 8.74TPTKK2480 pKa = 10.53LVADD2484 pKa = 5.11FFAFEE2489 pKa = 3.85PSLRR2493 pKa = 11.84NGAYY2497 pKa = 9.55VAAGDD2502 pKa = 4.18FNGDD2506 pKa = 3.09GYY2508 pKa = 11.81ADD2510 pKa = 4.2LVAGGGPDD2518 pKa = 3.31GAPRR2522 pKa = 11.84VEE2524 pKa = 4.3VLDD2527 pKa = 4.25GASLLSSNGQAPTMVANFFAGDD2549 pKa = 3.42PSLRR2553 pKa = 11.84SGARR2557 pKa = 11.84VAVKK2561 pKa = 10.56NLDD2564 pKa = 3.58GDD2566 pKa = 4.11GSADD2570 pKa = 3.72LVVGLATGTGTQVATYY2586 pKa = 10.0LGKK2589 pKa = 10.33DD2590 pKa = 3.41LTASGTPTAAAEE2602 pKa = 4.1LDD2604 pKa = 3.93PFPGFTGGVFVGG2616 pKa = 4.17
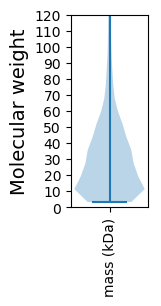
Molecular weight: 257.72 kDa
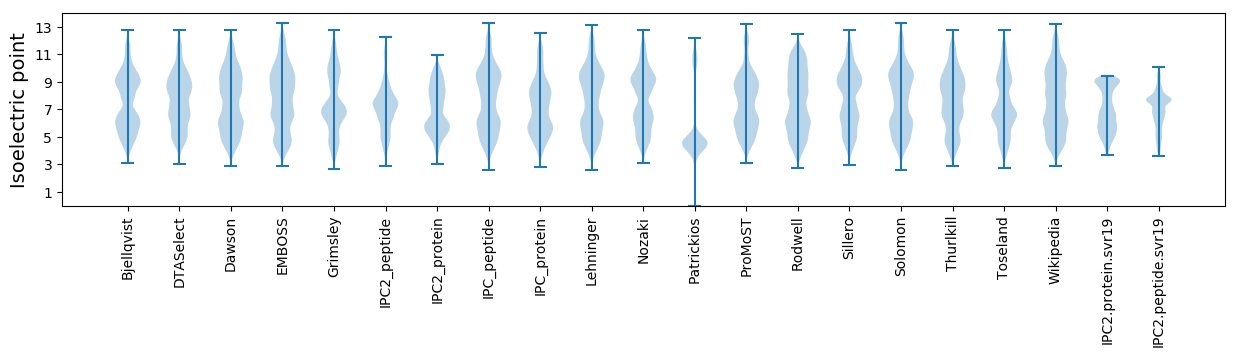
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A225DK67|A0A225DK67_9BACT Zinc protease OS=Fimbriiglobus ruber OX=1908690 GN=FRUB_06709 PE=4 SV=1
MM1 pKa = 7.5SFPRR5 pKa = 11.84KK6 pKa = 8.37FTGNLSVRR14 pKa = 11.84PVRR17 pKa = 11.84RR18 pKa = 11.84PAPGPRR24 pKa = 11.84WRR26 pKa = 11.84HH27 pKa = 4.8CWVTSWLGTGADD39 pKa = 3.47SRR41 pKa = 11.84RR42 pKa = 11.84VRR44 pKa = 11.84TRR46 pKa = 11.84RR47 pKa = 11.84PRR49 pKa = 11.84GTKK52 pKa = 10.06SPTALRR58 pKa = 11.84GSARR62 pKa = 11.84GNRR65 pKa = 11.84RR66 pKa = 11.84TARR69 pKa = 11.84PRR71 pKa = 11.84ASPAARR77 pKa = 11.84FVRR80 pKa = 11.84GGWW83 pKa = 3.16
MM1 pKa = 7.5SFPRR5 pKa = 11.84KK6 pKa = 8.37FTGNLSVRR14 pKa = 11.84PVRR17 pKa = 11.84RR18 pKa = 11.84PAPGPRR24 pKa = 11.84WRR26 pKa = 11.84HH27 pKa = 4.8CWVTSWLGTGADD39 pKa = 3.47SRR41 pKa = 11.84RR42 pKa = 11.84VRR44 pKa = 11.84TRR46 pKa = 11.84RR47 pKa = 11.84PRR49 pKa = 11.84GTKK52 pKa = 10.06SPTALRR58 pKa = 11.84GSARR62 pKa = 11.84GNRR65 pKa = 11.84RR66 pKa = 11.84TARR69 pKa = 11.84PRR71 pKa = 11.84ASPAARR77 pKa = 11.84FVRR80 pKa = 11.84GGWW83 pKa = 3.16
Molecular weight: 9.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3250331 |
37 |
8355 |
318.3 |
34.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.72 ± 0.04 | 0.981 ± 0.009 |
5.925 ± 0.02 | 5.172 ± 0.028 |
3.558 ± 0.017 | 8.793 ± 0.043 |
2.015 ± 0.015 | 3.848 ± 0.019 |
3.852 ± 0.032 | 9.365 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.712 ± 0.013 | 2.801 ± 0.024 |
6.283 ± 0.025 | 3.08 ± 0.016 |
7.132 ± 0.046 | 5.17 ± 0.036 |
6.565 ± 0.062 | 8.092 ± 0.024 |
1.524 ± 0.012 | 2.412 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
