
Erythrobacter longus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria;
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
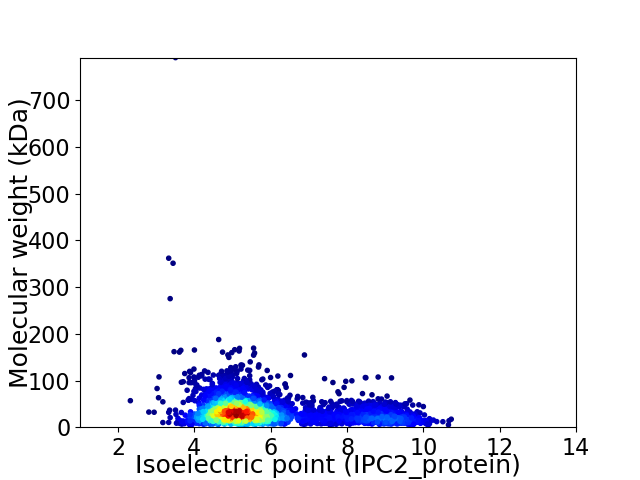
Virtual 2D-PAGE plot for 3219 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A074M7W5|A0A074M7W5_ERYLO Disulfide bond formation protein OS=Erythrobacter longus OX=1044 GN=EH31_12730 PE=4 SV=1
MM1 pKa = 7.25TKK3 pKa = 9.85IFRR6 pKa = 11.84TAAAGVALFAAVGLGSAAHH25 pKa = 5.76AQVTEE30 pKa = 4.32SADD33 pKa = 3.18ATAEE37 pKa = 4.04VLAALQLTNDD47 pKa = 3.5AGLDD51 pKa = 3.61FGAIVVNAGSTGGSVAVDD69 pKa = 3.5FTGFRR74 pKa = 11.84NCAGDD79 pKa = 4.01VICGPGGTEE88 pKa = 3.38QAAAFTVTGAAGTNVSVSVQDD109 pKa = 4.37LGANPVVLTHH119 pKa = 6.55TGFAGSTSNEE129 pKa = 3.78HH130 pKa = 6.8LIEE133 pKa = 4.24LTSLQDD139 pKa = 3.28GSFGGFPFFNGSEE152 pKa = 4.17TFNVGGTISLDD163 pKa = 3.25GTEE166 pKa = 3.82IAGTYY171 pKa = 9.25EE172 pKa = 3.84GSFDD176 pKa = 4.14VTVQYY181 pKa = 10.92QQ182 pKa = 2.97
MM1 pKa = 7.25TKK3 pKa = 9.85IFRR6 pKa = 11.84TAAAGVALFAAVGLGSAAHH25 pKa = 5.76AQVTEE30 pKa = 4.32SADD33 pKa = 3.18ATAEE37 pKa = 4.04VLAALQLTNDD47 pKa = 3.5AGLDD51 pKa = 3.61FGAIVVNAGSTGGSVAVDD69 pKa = 3.5FTGFRR74 pKa = 11.84NCAGDD79 pKa = 4.01VICGPGGTEE88 pKa = 3.38QAAAFTVTGAAGTNVSVSVQDD109 pKa = 4.37LGANPVVLTHH119 pKa = 6.55TGFAGSTSNEE129 pKa = 3.78HH130 pKa = 6.8LIEE133 pKa = 4.24LTSLQDD139 pKa = 3.28GSFGGFPFFNGSEE152 pKa = 4.17TFNVGGTISLDD163 pKa = 3.25GTEE166 pKa = 3.82IAGTYY171 pKa = 9.25EE172 pKa = 3.84GSFDD176 pKa = 4.14VTVQYY181 pKa = 10.92QQ182 pKa = 2.97
Molecular weight: 18.05 kDa
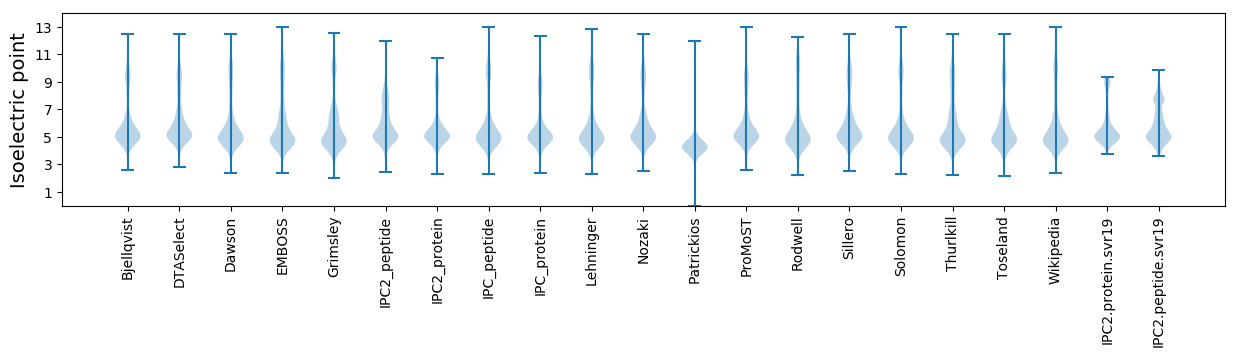
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A074M8S7|A0A074M8S7_ERYLO Short-chain dehydrogenase OS=Erythrobacter longus OX=1044 GN=EH31_00520 PE=3 SV=1
MM1 pKa = 7.05NAKK4 pKa = 10.05AGNEE8 pKa = 4.09PKK10 pKa = 10.68GSNAEE15 pKa = 3.86RR16 pKa = 11.84LALRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84MEE25 pKa = 4.0SGPAIFRR32 pKa = 11.84GAFRR36 pKa = 11.84PFFLGAAFWAIFTMCLWLGHH56 pKa = 5.9VLGQIPGGFVSDD68 pKa = 3.4ALAWHH73 pKa = 5.87RR74 pKa = 11.84HH75 pKa = 3.89EE76 pKa = 4.45MLFGFVGAAIAGFALTSAPNWTGRR100 pKa = 11.84LPIAGWPLATLFFFWCVGRR119 pKa = 11.84VLPFGLADD127 pKa = 3.92TSAALIALDD136 pKa = 3.36GGFYY140 pKa = 10.82LFLSFLIGRR149 pKa = 11.84EE150 pKa = 3.91EE151 pKa = 3.75THH153 pKa = 5.44GRR155 pKa = 11.84KK156 pKa = 9.22RR157 pKa = 11.84SLTICMVIAAFGFADD172 pKa = 3.03ILDD175 pKa = 4.0RR176 pKa = 11.84MEE178 pKa = 3.69MAKK181 pKa = 10.09WIDD184 pKa = 3.52SAGLGWRR191 pKa = 11.84AGIALVTALVMLIGGRR207 pKa = 11.84LIPSFTHH214 pKa = 5.64NWLLKK219 pKa = 8.67QVRR222 pKa = 11.84LDD224 pKa = 3.73PTPTQPDD231 pKa = 3.45LTDD234 pKa = 5.13AIVMAVTIGGLLVWILARR252 pKa = 11.84ASGASGTLLMLAGLANFARR271 pKa = 11.84LARR274 pKa = 11.84WQGWRR279 pKa = 11.84CLPCLPVFVLHH290 pKa = 7.37LGYY293 pKa = 10.41FWLALGLLLLGAAQFGWISEE313 pKa = 4.35SPAIHH318 pKa = 6.52ALSTGAMATMILGVMSRR335 pKa = 11.84TSLAMTARR343 pKa = 11.84EE344 pKa = 4.01PSFNLAIASACVLITLAAIARR365 pKa = 11.84VVAGLQIGQSFVLLVVAGFGWLGAFGLFLVVFGPILSSPRR405 pKa = 11.84KK406 pKa = 9.08GALGKK411 pKa = 10.34
MM1 pKa = 7.05NAKK4 pKa = 10.05AGNEE8 pKa = 4.09PKK10 pKa = 10.68GSNAEE15 pKa = 3.86RR16 pKa = 11.84LALRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84MEE25 pKa = 4.0SGPAIFRR32 pKa = 11.84GAFRR36 pKa = 11.84PFFLGAAFWAIFTMCLWLGHH56 pKa = 5.9VLGQIPGGFVSDD68 pKa = 3.4ALAWHH73 pKa = 5.87RR74 pKa = 11.84HH75 pKa = 3.89EE76 pKa = 4.45MLFGFVGAAIAGFALTSAPNWTGRR100 pKa = 11.84LPIAGWPLATLFFFWCVGRR119 pKa = 11.84VLPFGLADD127 pKa = 3.92TSAALIALDD136 pKa = 3.36GGFYY140 pKa = 10.82LFLSFLIGRR149 pKa = 11.84EE150 pKa = 3.91EE151 pKa = 3.75THH153 pKa = 5.44GRR155 pKa = 11.84KK156 pKa = 9.22RR157 pKa = 11.84SLTICMVIAAFGFADD172 pKa = 3.03ILDD175 pKa = 4.0RR176 pKa = 11.84MEE178 pKa = 3.69MAKK181 pKa = 10.09WIDD184 pKa = 3.52SAGLGWRR191 pKa = 11.84AGIALVTALVMLIGGRR207 pKa = 11.84LIPSFTHH214 pKa = 5.64NWLLKK219 pKa = 8.67QVRR222 pKa = 11.84LDD224 pKa = 3.73PTPTQPDD231 pKa = 3.45LTDD234 pKa = 5.13AIVMAVTIGGLLVWILARR252 pKa = 11.84ASGASGTLLMLAGLANFARR271 pKa = 11.84LARR274 pKa = 11.84WQGWRR279 pKa = 11.84CLPCLPVFVLHH290 pKa = 7.37LGYY293 pKa = 10.41FWLALGLLLLGAAQFGWISEE313 pKa = 4.35SPAIHH318 pKa = 6.52ALSTGAMATMILGVMSRR335 pKa = 11.84TSLAMTARR343 pKa = 11.84EE344 pKa = 4.01PSFNLAIASACVLITLAAIARR365 pKa = 11.84VVAGLQIGQSFVLLVVAGFGWLGAFGLFLVVFGPILSSPRR405 pKa = 11.84KK406 pKa = 9.08GALGKK411 pKa = 10.34
Molecular weight: 44.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1072962 |
41 |
7517 |
333.3 |
36.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.987 ± 0.059 | 0.807 ± 0.017 |
6.194 ± 0.064 | 6.495 ± 0.042 |
3.903 ± 0.034 | 8.816 ± 0.056 |
1.809 ± 0.027 | 5.287 ± 0.027 |
3.477 ± 0.049 | 9.539 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.444 ± 0.028 | 3.013 ± 0.035 |
4.859 ± 0.038 | 3.355 ± 0.025 |
6.33 ± 0.05 | 5.89 ± 0.037 |
5.37 ± 0.042 | 6.901 ± 0.033 |
1.351 ± 0.02 | 2.176 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
