
Paxillus involutus ATCC 200175
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Boletales; Paxilineae; Paxillaceae; Paxillus; Paxillus involutus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
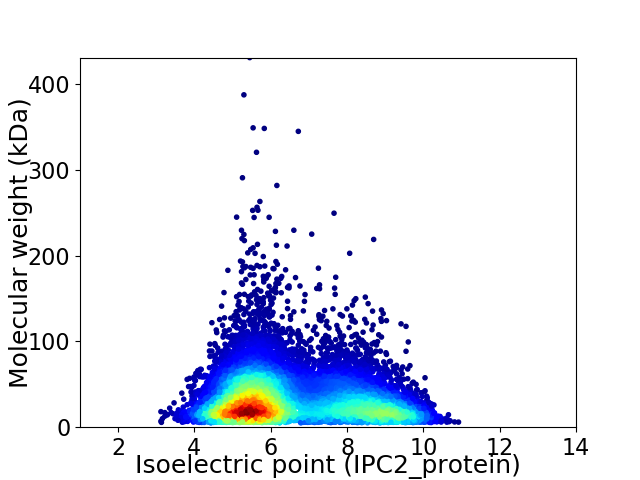
Virtual 2D-PAGE plot for 11689 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C9TB44|A0A0C9TB44_PAXIN Unplaced genomic scaffold PAXINscaffold_37 whole genome shotgun sequence OS=Paxillus involutus ATCC 200175 OX=664439 GN=PAXINDRAFT_156773 PE=4 SV=1
MM1 pKa = 7.43INVFTSLLDD10 pKa = 3.53VGPTAEE16 pKa = 4.51GEE18 pKa = 4.44IIAPMADD25 pKa = 3.08NLLAAGKK32 pKa = 7.26WLKK35 pKa = 10.55YY36 pKa = 10.4AGEE39 pKa = 4.34CVYY42 pKa = 10.45ATDD45 pKa = 3.29YY46 pKa = 10.46WYY48 pKa = 8.27QTSQDD53 pKa = 3.52PTGSFRR59 pKa = 11.84FLTTPQTFCIVAFNKK74 pKa = 7.34PTNGSVVVNAGGVVLPIQQGDD95 pKa = 4.38AIRR98 pKa = 11.84LLGPNSPGVFSDD110 pKa = 3.58DD111 pKa = 3.05TTAQTSGLEE120 pKa = 3.74WRR122 pKa = 11.84MDD124 pKa = 3.51EE125 pKa = 5.65DD126 pKa = 4.11GVLTIDD132 pKa = 3.83VPEE135 pKa = 4.58DD136 pKa = 3.19QVDD139 pKa = 3.72QVDD142 pKa = 4.18YY143 pKa = 10.82AWAFQVSYY151 pKa = 11.67ALFF154 pKa = 4.12
MM1 pKa = 7.43INVFTSLLDD10 pKa = 3.53VGPTAEE16 pKa = 4.51GEE18 pKa = 4.44IIAPMADD25 pKa = 3.08NLLAAGKK32 pKa = 7.26WLKK35 pKa = 10.55YY36 pKa = 10.4AGEE39 pKa = 4.34CVYY42 pKa = 10.45ATDD45 pKa = 3.29YY46 pKa = 10.46WYY48 pKa = 8.27QTSQDD53 pKa = 3.52PTGSFRR59 pKa = 11.84FLTTPQTFCIVAFNKK74 pKa = 7.34PTNGSVVVNAGGVVLPIQQGDD95 pKa = 4.38AIRR98 pKa = 11.84LLGPNSPGVFSDD110 pKa = 3.58DD111 pKa = 3.05TTAQTSGLEE120 pKa = 3.74WRR122 pKa = 11.84MDD124 pKa = 3.51EE125 pKa = 5.65DD126 pKa = 4.11GVLTIDD132 pKa = 3.83VPEE135 pKa = 4.58DD136 pKa = 3.19QVDD139 pKa = 3.72QVDD142 pKa = 4.18YY143 pKa = 10.82AWAFQVSYY151 pKa = 11.67ALFF154 pKa = 4.12
Molecular weight: 16.74 kDa
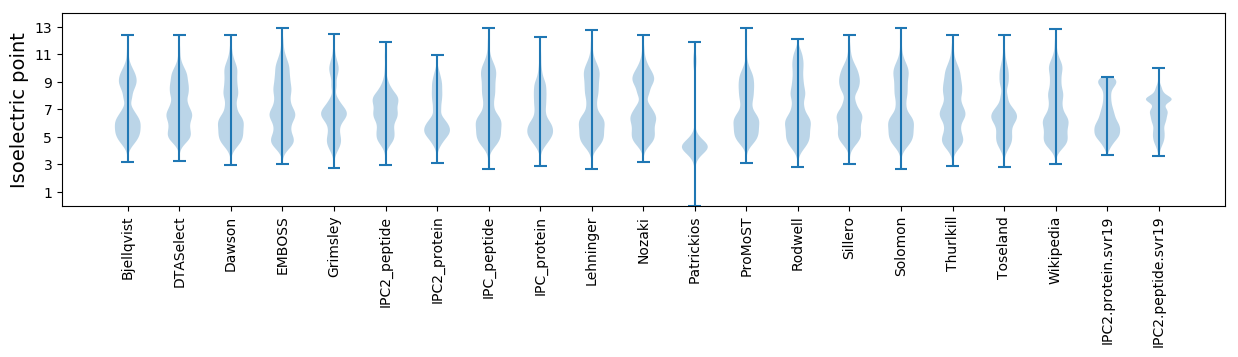
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C9SN70|A0A0C9SN70_PAXIN Unplaced genomic scaffold PAXINscaffold_580 whole genome shotgun sequence (Fragment) OS=Paxillus involutus ATCC 200175 OX=664439 GN=PAXINDRAFT_90432 PE=4 SV=1
MM1 pKa = 7.27AQHH4 pKa = 6.07FHH6 pKa = 6.16FALRR10 pKa = 11.84ALAYY14 pKa = 9.94VAISLCPCFRR24 pKa = 11.84QVIVTSPGVNGAVRR38 pKa = 11.84LLVNILGWNEE48 pKa = 4.03SLSLLSSRR56 pKa = 11.84FCNILEE62 pKa = 4.31AFHH65 pKa = 6.68SRR67 pKa = 11.84LHH69 pKa = 5.35IVARR73 pKa = 11.84QTQQNGVASVDD84 pKa = 3.38ASVDD88 pKa = 3.72SLHH91 pKa = 6.97LISLRR96 pKa = 11.84LILPRR101 pKa = 11.84YY102 pKa = 8.27LARR105 pKa = 11.84SPTQAHH111 pKa = 6.21RR112 pKa = 11.84AKK114 pKa = 10.72FSIPPFILVFGKK126 pKa = 8.83ATSTGVHH133 pKa = 5.97SPSCASYY140 pKa = 10.39HH141 pKa = 6.0RR142 pKa = 11.84HH143 pKa = 5.69TGPLKK148 pKa = 10.73DD149 pKa = 3.67VTLSASVLNTATYY162 pKa = 10.81CLARR166 pKa = 11.84PPKK169 pKa = 10.09QSPDD173 pKa = 3.01PSFGVFYY180 pKa = 10.6WEE182 pKa = 4.03TSLVHH187 pKa = 6.29LAGILIPGCRR197 pKa = 11.84SSSRR201 pKa = 11.84PPQHH205 pKa = 5.41WKK207 pKa = 9.03MLIVSQDD214 pKa = 3.24RR215 pKa = 11.84LIGGLTQTVSEE226 pKa = 4.71GARR229 pKa = 11.84TRR231 pKa = 11.84SPARR235 pKa = 11.84FMVDD239 pKa = 2.99AARR242 pKa = 11.84TEE244 pKa = 4.29TGATLPQYY252 pKa = 11.13KK253 pKa = 8.75STTEE257 pKa = 3.73AMSTSHH263 pKa = 6.56SPHH266 pKa = 7.5SMQTWHH272 pKa = 6.15SRR274 pKa = 11.84KK275 pKa = 10.2GSTSFPAISVPPTTVVAIMVHH296 pKa = 6.5PEE298 pKa = 4.0SLHH301 pKa = 6.53CLAQSRR307 pKa = 11.84FGYY310 pKa = 10.11LPPIRR315 pKa = 11.84IASRR319 pKa = 11.84RR320 pKa = 11.84ACPEE324 pKa = 3.74EE325 pKa = 4.03DD326 pKa = 3.5TGDD329 pKa = 3.71LTAVWAQEE337 pKa = 3.71WPAPSIGGRR346 pKa = 11.84PCRR349 pKa = 11.84NNGRR353 pKa = 11.84RR354 pKa = 11.84KK355 pKa = 9.37FLVVSRR361 pKa = 11.84RR362 pKa = 11.84MNLPLKK368 pKa = 10.11QAA370 pKa = 3.81
MM1 pKa = 7.27AQHH4 pKa = 6.07FHH6 pKa = 6.16FALRR10 pKa = 11.84ALAYY14 pKa = 9.94VAISLCPCFRR24 pKa = 11.84QVIVTSPGVNGAVRR38 pKa = 11.84LLVNILGWNEE48 pKa = 4.03SLSLLSSRR56 pKa = 11.84FCNILEE62 pKa = 4.31AFHH65 pKa = 6.68SRR67 pKa = 11.84LHH69 pKa = 5.35IVARR73 pKa = 11.84QTQQNGVASVDD84 pKa = 3.38ASVDD88 pKa = 3.72SLHH91 pKa = 6.97LISLRR96 pKa = 11.84LILPRR101 pKa = 11.84YY102 pKa = 8.27LARR105 pKa = 11.84SPTQAHH111 pKa = 6.21RR112 pKa = 11.84AKK114 pKa = 10.72FSIPPFILVFGKK126 pKa = 8.83ATSTGVHH133 pKa = 5.97SPSCASYY140 pKa = 10.39HH141 pKa = 6.0RR142 pKa = 11.84HH143 pKa = 5.69TGPLKK148 pKa = 10.73DD149 pKa = 3.67VTLSASVLNTATYY162 pKa = 10.81CLARR166 pKa = 11.84PPKK169 pKa = 10.09QSPDD173 pKa = 3.01PSFGVFYY180 pKa = 10.6WEE182 pKa = 4.03TSLVHH187 pKa = 6.29LAGILIPGCRR197 pKa = 11.84SSSRR201 pKa = 11.84PPQHH205 pKa = 5.41WKK207 pKa = 9.03MLIVSQDD214 pKa = 3.24RR215 pKa = 11.84LIGGLTQTVSEE226 pKa = 4.71GARR229 pKa = 11.84TRR231 pKa = 11.84SPARR235 pKa = 11.84FMVDD239 pKa = 2.99AARR242 pKa = 11.84TEE244 pKa = 4.29TGATLPQYY252 pKa = 11.13KK253 pKa = 8.75STTEE257 pKa = 3.73AMSTSHH263 pKa = 6.56SPHH266 pKa = 7.5SMQTWHH272 pKa = 6.15SRR274 pKa = 11.84KK275 pKa = 10.2GSTSFPAISVPPTTVVAIMVHH296 pKa = 6.5PEE298 pKa = 4.0SLHH301 pKa = 6.53CLAQSRR307 pKa = 11.84FGYY310 pKa = 10.11LPPIRR315 pKa = 11.84IASRR319 pKa = 11.84RR320 pKa = 11.84ACPEE324 pKa = 3.74EE325 pKa = 4.03DD326 pKa = 3.5TGDD329 pKa = 3.71LTAVWAQEE337 pKa = 3.71WPAPSIGGRR346 pKa = 11.84PCRR349 pKa = 11.84NNGRR353 pKa = 11.84RR354 pKa = 11.84KK355 pKa = 9.37FLVVSRR361 pKa = 11.84RR362 pKa = 11.84MNLPLKK368 pKa = 10.11QAA370 pKa = 3.81
Molecular weight: 40.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3729177 |
49 |
3922 |
319.0 |
35.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.115 ± 0.023 | 1.523 ± 0.012 |
5.634 ± 0.019 | 5.898 ± 0.023 |
3.657 ± 0.016 | 6.471 ± 0.02 |
2.965 ± 0.013 | 4.856 ± 0.016 |
4.434 ± 0.019 | 9.165 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.14 ± 0.01 | 3.46 ± 0.013 |
6.439 ± 0.031 | 3.862 ± 0.014 |
6.13 ± 0.024 | 8.422 ± 0.03 |
6.18 ± 0.018 | 6.471 ± 0.016 |
1.604 ± 0.012 | 2.576 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
