
Cimodo virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; unclassified Reoviridae
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
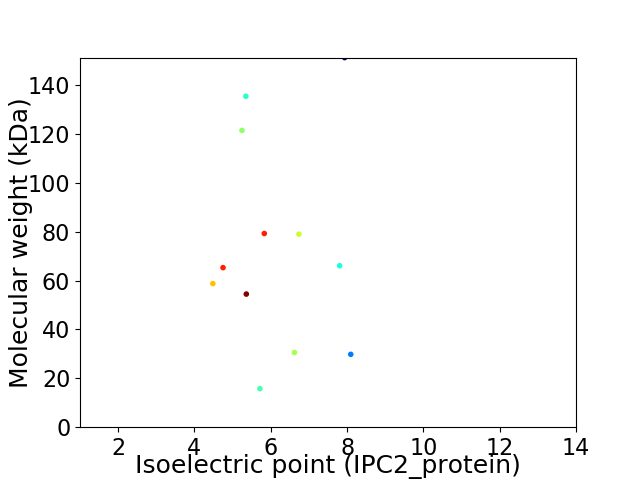
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5RUY0|W5RUY0_9REOV Uncharacterized protein OS=Cimodo virus OX=1427476 PE=4 SV=1
MM1 pKa = 7.6AAQPNNYY8 pKa = 9.39DD9 pKa = 3.29PQFFRR14 pKa = 11.84QATLLVDD21 pKa = 3.98TIRR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 9.06ATDD29 pKa = 4.07KK30 pKa = 11.27FNFDD34 pKa = 3.34NARR37 pKa = 11.84NPVYY41 pKa = 9.81PANYY45 pKa = 9.49NLRR48 pKa = 11.84LGYY51 pKa = 9.48MPARR55 pKa = 11.84TSFPVFFDD63 pKa = 3.89TYY65 pKa = 10.32QPPIQSPRR73 pKa = 11.84EE74 pKa = 3.91YY75 pKa = 10.58LDD77 pKa = 3.05TFGPVQYY84 pKa = 9.52TVNDD88 pKa = 3.92ALTDD92 pKa = 3.64DD93 pKa = 4.55EE94 pKa = 4.71KK95 pKa = 11.39RR96 pKa = 11.84QVFQNIGDD104 pKa = 4.54LLQQIANAYY113 pKa = 8.71HH114 pKa = 6.04LQLNVEE120 pKa = 4.4QIDD123 pKa = 3.77VTEE126 pKa = 4.24TVPQLVYY133 pKa = 9.88TAFLLPDD140 pKa = 3.44GTVDD144 pKa = 3.31QDD146 pKa = 3.51YY147 pKa = 11.14ARR149 pKa = 11.84YY150 pKa = 7.77MQSTNPQNQDD160 pKa = 2.52VAQFEE165 pKa = 5.04LIRR168 pKa = 11.84ANGYY172 pKa = 10.35GGFEE176 pKa = 4.1FRR178 pKa = 11.84DD179 pKa = 3.53PAEE182 pKa = 4.55DD183 pKa = 3.42VLHH186 pKa = 7.04RR187 pKa = 11.84EE188 pKa = 4.45DD189 pKa = 4.34GNVIEE194 pKa = 4.31PAPRR198 pKa = 11.84RR199 pKa = 11.84RR200 pKa = 11.84IIDD203 pKa = 3.75PASDD207 pKa = 3.88DD208 pKa = 3.91SDD210 pKa = 3.9VDD212 pKa = 3.6EE213 pKa = 6.33DD214 pKa = 5.04DD215 pKa = 4.46VPEE218 pKa = 3.97QEE220 pKa = 4.1RR221 pKa = 11.84RR222 pKa = 11.84FAAEE226 pKa = 3.66QNRR229 pKa = 11.84QQSVTILKK237 pKa = 10.32GRR239 pKa = 11.84VIPIDD244 pKa = 3.61ALRR247 pKa = 11.84VRR249 pKa = 11.84CWYY252 pKa = 9.07EE253 pKa = 4.12GPAQIPDD260 pKa = 4.74AIDD263 pKa = 3.77FPAISDD269 pKa = 4.07EE270 pKa = 5.48IRR272 pKa = 11.84IQDD275 pKa = 3.49NSSSFDD281 pKa = 3.37FFACVSNAMMNGAFQDD297 pKa = 3.65YY298 pKa = 10.02NYY300 pKa = 11.22KK301 pKa = 10.44SFCTMLVNYY310 pKa = 9.24CWDD313 pKa = 3.46YY314 pKa = 11.46AISDD318 pKa = 4.06RR319 pKa = 11.84IGYY322 pKa = 9.23FVMRR326 pKa = 11.84MTKK329 pKa = 9.96YY330 pKa = 10.37PVGQVQAPRR339 pKa = 11.84EE340 pKa = 4.15VVATRR345 pKa = 11.84EE346 pKa = 3.86QLEE349 pKa = 4.18AKK351 pKa = 10.2PNGEE355 pKa = 5.28FYY357 pKa = 9.99WQMYY361 pKa = 9.42RR362 pKa = 11.84YY363 pKa = 6.87WTITKK368 pKa = 10.08RR369 pKa = 11.84LNDD372 pKa = 3.39EE373 pKa = 4.23ALASFVQPSINILANEE389 pKa = 3.98IGLFEE394 pKa = 4.74GGPGNVLNATQNIKK408 pKa = 10.41FQMEE412 pKa = 4.18DD413 pKa = 3.07RR414 pKa = 11.84QINYY418 pKa = 8.74PDD420 pKa = 4.47LRR422 pKa = 11.84LACPSLDD429 pKa = 3.67YY430 pKa = 10.78EE431 pKa = 4.36QLVLFPADD439 pKa = 2.99SFAVMRR445 pKa = 11.84VLVSAIMVTTANKK458 pKa = 9.94RR459 pKa = 11.84DD460 pKa = 3.76VLIDD464 pKa = 3.5PVNQARR470 pKa = 11.84YY471 pKa = 10.1NEE473 pKa = 3.87WTLYY477 pKa = 10.94DD478 pKa = 3.71MKK480 pKa = 10.98EE481 pKa = 4.09VTTKK485 pKa = 10.8SSLRR489 pKa = 11.84MKK491 pKa = 9.63WRR493 pKa = 11.84TSALICCMLLYY504 pKa = 10.66LLPDD508 pKa = 4.03DD509 pKa = 4.63
MM1 pKa = 7.6AAQPNNYY8 pKa = 9.39DD9 pKa = 3.29PQFFRR14 pKa = 11.84QATLLVDD21 pKa = 3.98TIRR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 9.06ATDD29 pKa = 4.07KK30 pKa = 11.27FNFDD34 pKa = 3.34NARR37 pKa = 11.84NPVYY41 pKa = 9.81PANYY45 pKa = 9.49NLRR48 pKa = 11.84LGYY51 pKa = 9.48MPARR55 pKa = 11.84TSFPVFFDD63 pKa = 3.89TYY65 pKa = 10.32QPPIQSPRR73 pKa = 11.84EE74 pKa = 3.91YY75 pKa = 10.58LDD77 pKa = 3.05TFGPVQYY84 pKa = 9.52TVNDD88 pKa = 3.92ALTDD92 pKa = 3.64DD93 pKa = 4.55EE94 pKa = 4.71KK95 pKa = 11.39RR96 pKa = 11.84QVFQNIGDD104 pKa = 4.54LLQQIANAYY113 pKa = 8.71HH114 pKa = 6.04LQLNVEE120 pKa = 4.4QIDD123 pKa = 3.77VTEE126 pKa = 4.24TVPQLVYY133 pKa = 9.88TAFLLPDD140 pKa = 3.44GTVDD144 pKa = 3.31QDD146 pKa = 3.51YY147 pKa = 11.14ARR149 pKa = 11.84YY150 pKa = 7.77MQSTNPQNQDD160 pKa = 2.52VAQFEE165 pKa = 5.04LIRR168 pKa = 11.84ANGYY172 pKa = 10.35GGFEE176 pKa = 4.1FRR178 pKa = 11.84DD179 pKa = 3.53PAEE182 pKa = 4.55DD183 pKa = 3.42VLHH186 pKa = 7.04RR187 pKa = 11.84EE188 pKa = 4.45DD189 pKa = 4.34GNVIEE194 pKa = 4.31PAPRR198 pKa = 11.84RR199 pKa = 11.84RR200 pKa = 11.84IIDD203 pKa = 3.75PASDD207 pKa = 3.88DD208 pKa = 3.91SDD210 pKa = 3.9VDD212 pKa = 3.6EE213 pKa = 6.33DD214 pKa = 5.04DD215 pKa = 4.46VPEE218 pKa = 3.97QEE220 pKa = 4.1RR221 pKa = 11.84RR222 pKa = 11.84FAAEE226 pKa = 3.66QNRR229 pKa = 11.84QQSVTILKK237 pKa = 10.32GRR239 pKa = 11.84VIPIDD244 pKa = 3.61ALRR247 pKa = 11.84VRR249 pKa = 11.84CWYY252 pKa = 9.07EE253 pKa = 4.12GPAQIPDD260 pKa = 4.74AIDD263 pKa = 3.77FPAISDD269 pKa = 4.07EE270 pKa = 5.48IRR272 pKa = 11.84IQDD275 pKa = 3.49NSSSFDD281 pKa = 3.37FFACVSNAMMNGAFQDD297 pKa = 3.65YY298 pKa = 10.02NYY300 pKa = 11.22KK301 pKa = 10.44SFCTMLVNYY310 pKa = 9.24CWDD313 pKa = 3.46YY314 pKa = 11.46AISDD318 pKa = 4.06RR319 pKa = 11.84IGYY322 pKa = 9.23FVMRR326 pKa = 11.84MTKK329 pKa = 9.96YY330 pKa = 10.37PVGQVQAPRR339 pKa = 11.84EE340 pKa = 4.15VVATRR345 pKa = 11.84EE346 pKa = 3.86QLEE349 pKa = 4.18AKK351 pKa = 10.2PNGEE355 pKa = 5.28FYY357 pKa = 9.99WQMYY361 pKa = 9.42RR362 pKa = 11.84YY363 pKa = 6.87WTITKK368 pKa = 10.08RR369 pKa = 11.84LNDD372 pKa = 3.39EE373 pKa = 4.23ALASFVQPSINILANEE389 pKa = 3.98IGLFEE394 pKa = 4.74GGPGNVLNATQNIKK408 pKa = 10.41FQMEE412 pKa = 4.18DD413 pKa = 3.07RR414 pKa = 11.84QINYY418 pKa = 8.74PDD420 pKa = 4.47LRR422 pKa = 11.84LACPSLDD429 pKa = 3.67YY430 pKa = 10.78EE431 pKa = 4.36QLVLFPADD439 pKa = 2.99SFAVMRR445 pKa = 11.84VLVSAIMVTTANKK458 pKa = 9.94RR459 pKa = 11.84DD460 pKa = 3.76VLIDD464 pKa = 3.5PVNQARR470 pKa = 11.84YY471 pKa = 10.1NEE473 pKa = 3.87WTLYY477 pKa = 10.94DD478 pKa = 3.71MKK480 pKa = 10.98EE481 pKa = 4.09VTTKK485 pKa = 10.8SSLRR489 pKa = 11.84MKK491 pKa = 9.63WRR493 pKa = 11.84TSALICCMLLYY504 pKa = 10.66LLPDD508 pKa = 4.03DD509 pKa = 4.63
Molecular weight: 58.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5RUY0|W5RUY0_9REOV Uncharacterized protein OS=Cimodo virus OX=1427476 PE=4 SV=1
MM1 pKa = 7.19ATIEE5 pKa = 4.46IPFSQATQYY14 pKa = 10.91EE15 pKa = 4.61HH16 pKa = 7.04IANKK20 pKa = 10.13RR21 pKa = 11.84NHH23 pKa = 5.62QLVNIIMEE31 pKa = 4.69ANQFLLRR38 pKa = 11.84HH39 pKa = 5.12QFVLSVNYY47 pKa = 9.54FGPKK51 pKa = 9.37EE52 pKa = 4.0YY53 pKa = 10.58EE54 pKa = 4.03PNAILHH60 pKa = 5.63PCYY63 pKa = 10.29FRR65 pKa = 11.84FALYY69 pKa = 10.57NNRR72 pKa = 11.84ASIPCRR78 pKa = 11.84TRR80 pKa = 11.84YY81 pKa = 9.89LYY83 pKa = 11.04LSLPDD88 pKa = 3.23SSGPINSDD96 pKa = 3.25SQKK99 pKa = 9.91SAEE102 pKa = 4.61SIRR105 pKa = 11.84KK106 pKa = 8.74SAFSRR111 pKa = 11.84IYY113 pKa = 8.94SQAVRR118 pKa = 11.84RR119 pKa = 11.84AVDD122 pKa = 4.47AIRR125 pKa = 11.84RR126 pKa = 11.84GEE128 pKa = 3.96LSTSVEE134 pKa = 4.03NVEE137 pKa = 4.26YY138 pKa = 10.18TSKK141 pKa = 9.94CTLKK145 pKa = 10.0RR146 pKa = 11.84TFEE149 pKa = 4.5SIAEE153 pKa = 4.17YY154 pKa = 10.8VPPTPFILRR163 pKa = 11.84YY164 pKa = 9.75CEE166 pKa = 3.86LFNAIFTLNLDD177 pKa = 3.55PKK179 pKa = 10.19EE180 pKa = 4.16VNEE183 pKa = 4.21EE184 pKa = 3.92GAKK187 pKa = 10.0RR188 pKa = 11.84EE189 pKa = 3.96QGKK192 pKa = 9.54RR193 pKa = 11.84CSLDD197 pKa = 3.12YY198 pKa = 11.3YY199 pKa = 10.42IGLKK203 pKa = 9.46YY204 pKa = 10.3RR205 pKa = 11.84WGRR208 pKa = 11.84PARR211 pKa = 11.84DD212 pKa = 4.29DD213 pKa = 3.01LWAYY217 pKa = 8.16QWEE220 pKa = 4.66RR221 pKa = 11.84YY222 pKa = 8.95ALNPDD227 pKa = 3.72PNDD230 pKa = 3.39PQKK233 pKa = 10.64ILRR236 pKa = 11.84EE237 pKa = 3.98HH238 pKa = 6.76RR239 pKa = 11.84EE240 pKa = 3.82GVEE243 pKa = 4.11DD244 pKa = 3.62VSRR247 pKa = 11.84TNKK250 pKa = 8.15ITFANN255 pKa = 3.77
MM1 pKa = 7.19ATIEE5 pKa = 4.46IPFSQATQYY14 pKa = 10.91EE15 pKa = 4.61HH16 pKa = 7.04IANKK20 pKa = 10.13RR21 pKa = 11.84NHH23 pKa = 5.62QLVNIIMEE31 pKa = 4.69ANQFLLRR38 pKa = 11.84HH39 pKa = 5.12QFVLSVNYY47 pKa = 9.54FGPKK51 pKa = 9.37EE52 pKa = 4.0YY53 pKa = 10.58EE54 pKa = 4.03PNAILHH60 pKa = 5.63PCYY63 pKa = 10.29FRR65 pKa = 11.84FALYY69 pKa = 10.57NNRR72 pKa = 11.84ASIPCRR78 pKa = 11.84TRR80 pKa = 11.84YY81 pKa = 9.89LYY83 pKa = 11.04LSLPDD88 pKa = 3.23SSGPINSDD96 pKa = 3.25SQKK99 pKa = 9.91SAEE102 pKa = 4.61SIRR105 pKa = 11.84KK106 pKa = 8.74SAFSRR111 pKa = 11.84IYY113 pKa = 8.94SQAVRR118 pKa = 11.84RR119 pKa = 11.84AVDD122 pKa = 4.47AIRR125 pKa = 11.84RR126 pKa = 11.84GEE128 pKa = 3.96LSTSVEE134 pKa = 4.03NVEE137 pKa = 4.26YY138 pKa = 10.18TSKK141 pKa = 9.94CTLKK145 pKa = 10.0RR146 pKa = 11.84TFEE149 pKa = 4.5SIAEE153 pKa = 4.17YY154 pKa = 10.8VPPTPFILRR163 pKa = 11.84YY164 pKa = 9.75CEE166 pKa = 3.86LFNAIFTLNLDD177 pKa = 3.55PKK179 pKa = 10.19EE180 pKa = 4.16VNEE183 pKa = 4.21EE184 pKa = 3.92GAKK187 pKa = 10.0RR188 pKa = 11.84EE189 pKa = 3.96QGKK192 pKa = 9.54RR193 pKa = 11.84CSLDD197 pKa = 3.12YY198 pKa = 11.3YY199 pKa = 10.42IGLKK203 pKa = 9.46YY204 pKa = 10.3RR205 pKa = 11.84WGRR208 pKa = 11.84PARR211 pKa = 11.84DD212 pKa = 4.29DD213 pKa = 3.01LWAYY217 pKa = 8.16QWEE220 pKa = 4.66RR221 pKa = 11.84YY222 pKa = 8.95ALNPDD227 pKa = 3.72PNDD230 pKa = 3.39PQKK233 pKa = 10.64ILRR236 pKa = 11.84EE237 pKa = 3.98HH238 pKa = 6.76RR239 pKa = 11.84EE240 pKa = 3.82GVEE243 pKa = 4.11DD244 pKa = 3.62VSRR247 pKa = 11.84TNKK250 pKa = 8.15ITFANN255 pKa = 3.77
Molecular weight: 29.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7859 |
140 |
1334 |
654.9 |
73.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.647 ± 0.364 | 0.98 ± 0.142 |
6.006 ± 0.286 | 5.382 ± 0.202 |
4.479 ± 0.167 | 4.899 ± 0.229 |
2.239 ± 0.3 | 6.438 ± 0.225 |
4.314 ± 0.4 | 8.932 ± 0.372 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.227 ± 0.179 | 4.708 ± 0.274 |
5.535 ± 0.298 | 4.632 ± 0.263 |
5.84 ± 0.319 | 7.635 ± 0.605 |
6.235 ± 0.354 | 6.489 ± 0.254 |
1.107 ± 0.083 | 4.275 ± 0.313 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
