
Nocardioides sp. S-1144
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
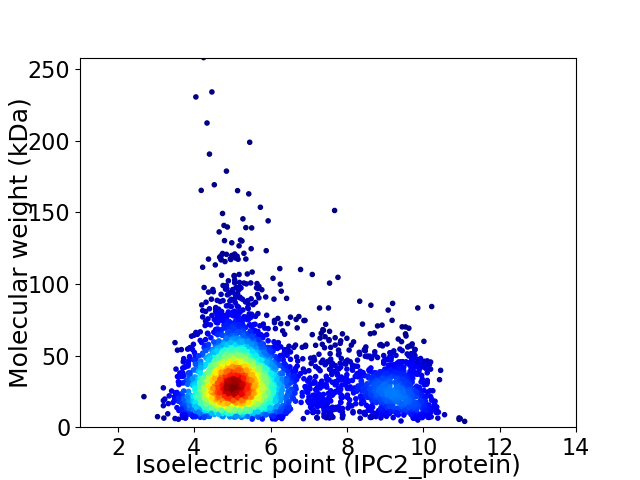
Virtual 2D-PAGE plot for 4010 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B7REY2|A0A5B7REY2_9ACTN Uncharacterized protein OS=Nocardioides sp. S-1144 OX=2582905 GN=FE634_12710 PE=4 SV=1
MM1 pKa = 6.65TRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84QRR7 pKa = 11.84VLAGTTLVSLALLAAGCGTAGKK29 pKa = 9.58IATSSEE35 pKa = 4.27PSSSAAPSSPGSSSAGEE52 pKa = 4.68DD53 pKa = 3.45PTDD56 pKa = 4.02PAPEE60 pKa = 4.78ADD62 pKa = 4.67LDD64 pKa = 4.22LALSEE69 pKa = 4.3PAEE72 pKa = 4.09DD73 pKa = 3.13SYY75 pKa = 11.8YY76 pKa = 10.44PEE78 pKa = 4.82VGDD81 pKa = 4.26PSVDD85 pKa = 3.09ALRR88 pKa = 11.84YY89 pKa = 10.23DD90 pKa = 5.85LDD92 pKa = 4.26LTWDD96 pKa = 3.97PATDD100 pKa = 3.43TLTGTEE106 pKa = 4.21TLTFRR111 pKa = 11.84STGTAEE117 pKa = 3.96EE118 pKa = 4.22FQLDD122 pKa = 3.66FAEE125 pKa = 4.46PLEE128 pKa = 4.43ITSLTLDD135 pKa = 3.65GAEE138 pKa = 3.99VDD140 pKa = 3.63HH141 pKa = 5.95EE142 pKa = 4.51HH143 pKa = 7.02RR144 pKa = 11.84GKK146 pKa = 10.54DD147 pKa = 3.69LIVSAPVEE155 pKa = 3.63EE156 pKa = 4.68DD157 pKa = 3.01ARR159 pKa = 11.84HH160 pKa = 5.34EE161 pKa = 4.08LVVEE165 pKa = 4.29YY166 pKa = 10.3SGTPAPVPAPTEE178 pKa = 4.0RR179 pKa = 11.84SDD181 pKa = 3.89LATTGFTIDD190 pKa = 5.12AEE192 pKa = 4.43HH193 pKa = 5.71QTWTMQEE200 pKa = 3.84PFGAYY205 pKa = 6.78TWYY208 pKa = 10.72AVNDD212 pKa = 3.76QPADD216 pKa = 3.3KK217 pKa = 10.71ALYY220 pKa = 10.37DD221 pKa = 3.95FTLHH225 pKa = 6.98VPDD228 pKa = 4.53PWTGVANGAKK238 pKa = 10.27VSDD241 pKa = 3.65EE242 pKa = 4.37TADD245 pKa = 3.87GTHH248 pKa = 5.52TTAFHH253 pKa = 6.31LAEE256 pKa = 4.36PAAAYY261 pKa = 9.92LVTLAFGDD269 pKa = 3.94YY270 pKa = 10.83RR271 pKa = 11.84EE272 pKa = 4.43TTDD275 pKa = 3.37VGPNGVPITYY285 pKa = 7.47WTPRR289 pKa = 11.84DD290 pKa = 3.78MEE292 pKa = 5.05PGSGLRR298 pKa = 11.84YY299 pKa = 9.91SPEE302 pKa = 3.32AMAWLEE308 pKa = 4.45TYY310 pKa = 10.54LGPYY314 pKa = 9.74PFDD317 pKa = 3.23TAGILVVDD325 pKa = 4.67SEE327 pKa = 5.18SGMEE331 pKa = 4.12TQTMITLGDD340 pKa = 3.79TEE342 pKa = 4.63YY343 pKa = 11.86SMAEE347 pKa = 3.69NTVVHH352 pKa = 6.45EE353 pKa = 5.48FAHH356 pKa = 5.24QWYY359 pKa = 8.92GDD361 pKa = 3.69QVTPDD366 pKa = 3.35DD367 pKa = 4.7WRR369 pKa = 11.84DD370 pKa = 2.74VWMNEE375 pKa = 3.02GMAMYY380 pKa = 9.61LQLMWEE386 pKa = 4.64AEE388 pKa = 4.18QSDD391 pKa = 4.58VPLDD395 pKa = 3.84SYY397 pKa = 11.93LEE399 pKa = 4.69GYY401 pKa = 9.92AGYY404 pKa = 10.38EE405 pKa = 3.48SDD407 pKa = 4.58YY408 pKa = 10.34RR409 pKa = 11.84AQSGPPADD417 pKa = 4.19YY418 pKa = 11.17DD419 pKa = 3.64PDD421 pKa = 4.02AFGSSNVYY429 pKa = 10.11FSGAFFWHH437 pKa = 7.34ALRR440 pKa = 11.84EE441 pKa = 4.25QVGDD445 pKa = 3.65EE446 pKa = 4.3VFFAAVKK453 pKa = 9.9GWPQSQDD460 pKa = 2.74NRR462 pKa = 11.84STDD465 pKa = 3.34RR466 pKa = 11.84EE467 pKa = 4.09TLLAYY472 pKa = 10.74LEE474 pKa = 4.7DD475 pKa = 3.84QTGEE479 pKa = 4.12DD480 pKa = 6.14LDD482 pKa = 4.32VLWDD486 pKa = 3.32AWLLGDD492 pKa = 4.96TSPEE496 pKa = 4.02LPP498 pKa = 3.96
MM1 pKa = 6.65TRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84QRR7 pKa = 11.84VLAGTTLVSLALLAAGCGTAGKK29 pKa = 9.58IATSSEE35 pKa = 4.27PSSSAAPSSPGSSSAGEE52 pKa = 4.68DD53 pKa = 3.45PTDD56 pKa = 4.02PAPEE60 pKa = 4.78ADD62 pKa = 4.67LDD64 pKa = 4.22LALSEE69 pKa = 4.3PAEE72 pKa = 4.09DD73 pKa = 3.13SYY75 pKa = 11.8YY76 pKa = 10.44PEE78 pKa = 4.82VGDD81 pKa = 4.26PSVDD85 pKa = 3.09ALRR88 pKa = 11.84YY89 pKa = 10.23DD90 pKa = 5.85LDD92 pKa = 4.26LTWDD96 pKa = 3.97PATDD100 pKa = 3.43TLTGTEE106 pKa = 4.21TLTFRR111 pKa = 11.84STGTAEE117 pKa = 3.96EE118 pKa = 4.22FQLDD122 pKa = 3.66FAEE125 pKa = 4.46PLEE128 pKa = 4.43ITSLTLDD135 pKa = 3.65GAEE138 pKa = 3.99VDD140 pKa = 3.63HH141 pKa = 5.95EE142 pKa = 4.51HH143 pKa = 7.02RR144 pKa = 11.84GKK146 pKa = 10.54DD147 pKa = 3.69LIVSAPVEE155 pKa = 3.63EE156 pKa = 4.68DD157 pKa = 3.01ARR159 pKa = 11.84HH160 pKa = 5.34EE161 pKa = 4.08LVVEE165 pKa = 4.29YY166 pKa = 10.3SGTPAPVPAPTEE178 pKa = 4.0RR179 pKa = 11.84SDD181 pKa = 3.89LATTGFTIDD190 pKa = 5.12AEE192 pKa = 4.43HH193 pKa = 5.71QTWTMQEE200 pKa = 3.84PFGAYY205 pKa = 6.78TWYY208 pKa = 10.72AVNDD212 pKa = 3.76QPADD216 pKa = 3.3KK217 pKa = 10.71ALYY220 pKa = 10.37DD221 pKa = 3.95FTLHH225 pKa = 6.98VPDD228 pKa = 4.53PWTGVANGAKK238 pKa = 10.27VSDD241 pKa = 3.65EE242 pKa = 4.37TADD245 pKa = 3.87GTHH248 pKa = 5.52TTAFHH253 pKa = 6.31LAEE256 pKa = 4.36PAAAYY261 pKa = 9.92LVTLAFGDD269 pKa = 3.94YY270 pKa = 10.83RR271 pKa = 11.84EE272 pKa = 4.43TTDD275 pKa = 3.37VGPNGVPITYY285 pKa = 7.47WTPRR289 pKa = 11.84DD290 pKa = 3.78MEE292 pKa = 5.05PGSGLRR298 pKa = 11.84YY299 pKa = 9.91SPEE302 pKa = 3.32AMAWLEE308 pKa = 4.45TYY310 pKa = 10.54LGPYY314 pKa = 9.74PFDD317 pKa = 3.23TAGILVVDD325 pKa = 4.67SEE327 pKa = 5.18SGMEE331 pKa = 4.12TQTMITLGDD340 pKa = 3.79TEE342 pKa = 4.63YY343 pKa = 11.86SMAEE347 pKa = 3.69NTVVHH352 pKa = 6.45EE353 pKa = 5.48FAHH356 pKa = 5.24QWYY359 pKa = 8.92GDD361 pKa = 3.69QVTPDD366 pKa = 3.35DD367 pKa = 4.7WRR369 pKa = 11.84DD370 pKa = 2.74VWMNEE375 pKa = 3.02GMAMYY380 pKa = 9.61LQLMWEE386 pKa = 4.64AEE388 pKa = 4.18QSDD391 pKa = 4.58VPLDD395 pKa = 3.84SYY397 pKa = 11.93LEE399 pKa = 4.69GYY401 pKa = 9.92AGYY404 pKa = 10.38EE405 pKa = 3.48SDD407 pKa = 4.58YY408 pKa = 10.34RR409 pKa = 11.84AQSGPPADD417 pKa = 4.19YY418 pKa = 11.17DD419 pKa = 3.64PDD421 pKa = 4.02AFGSSNVYY429 pKa = 10.11FSGAFFWHH437 pKa = 7.34ALRR440 pKa = 11.84EE441 pKa = 4.25QVGDD445 pKa = 3.65EE446 pKa = 4.3VFFAAVKK453 pKa = 9.9GWPQSQDD460 pKa = 2.74NRR462 pKa = 11.84STDD465 pKa = 3.34RR466 pKa = 11.84EE467 pKa = 4.09TLLAYY472 pKa = 10.74LEE474 pKa = 4.7DD475 pKa = 3.84QTGEE479 pKa = 4.12DD480 pKa = 6.14LDD482 pKa = 4.32VLWDD486 pKa = 3.32AWLLGDD492 pKa = 4.96TSPEE496 pKa = 4.02LPP498 pKa = 3.96
Molecular weight: 54.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B7RBN2|A0A5B7RBN2_9ACTN 30S ribosomal protein S14 type Z OS=Nocardioides sp. S-1144 OX=2582905 GN=rpsZ PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.24TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.24TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.07LGKK33 pKa = 9.87
Molecular weight: 4.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1318656 |
33 |
2466 |
328.8 |
35.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.433 ± 0.048 | 0.708 ± 0.01 |
6.799 ± 0.03 | 5.567 ± 0.033 |
2.627 ± 0.022 | 9.522 ± 0.035 |
2.227 ± 0.019 | 2.927 ± 0.029 |
1.613 ± 0.028 | 10.337 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.655 ± 0.015 | 1.589 ± 0.022 |
5.856 ± 0.032 | 2.544 ± 0.016 |
7.974 ± 0.045 | 5.028 ± 0.03 |
6.255 ± 0.04 | 10.036 ± 0.042 |
1.474 ± 0.016 | 1.829 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
