
False black widow spider associated circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.62
Get precalculated fractions of proteins
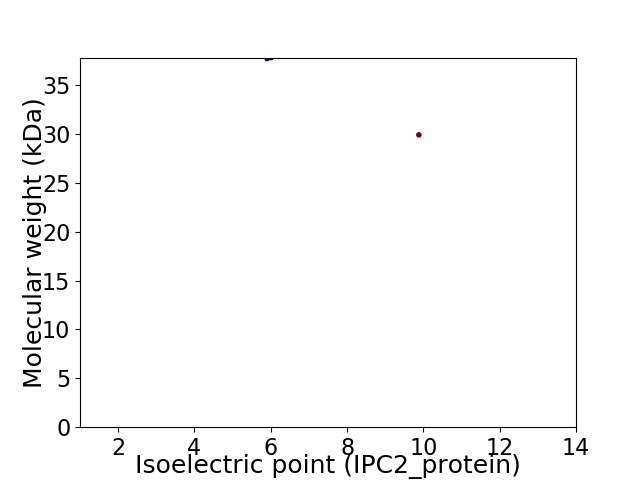
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A346BPE8|A0A346BPE8_9VIRU Putative capsid protein OS=False black widow spider associated circular virus 1 OX=2293279 PE=4 SV=1
MM1 pKa = 7.57SRR3 pKa = 11.84GRR5 pKa = 11.84NWLFTEE11 pKa = 4.47NNPTLNPNDD20 pKa = 3.66FLEE23 pKa = 4.52SAKK26 pKa = 10.71LWDD29 pKa = 3.54HH30 pKa = 5.18VRR32 pKa = 11.84YY33 pKa = 10.0IIFQQEE39 pKa = 3.93TGEE42 pKa = 4.75GGTSHH47 pKa = 5.67YY48 pKa = 10.16QGYY51 pKa = 9.47VEE53 pKa = 4.96FSQPIRR59 pKa = 11.84FAKK62 pKa = 8.92VTGFLPRR69 pKa = 11.84AHH71 pKa = 6.55WEE73 pKa = 3.86MRR75 pKa = 11.84RR76 pKa = 11.84GTSAQCIEE84 pKa = 4.5YY85 pKa = 10.29CSKK88 pKa = 11.12LDD90 pKa = 3.65TRR92 pKa = 11.84VDD94 pKa = 4.17GPWTYY99 pKa = 11.41GDD101 pKa = 4.52PGFSSQGSRR110 pKa = 11.84SDD112 pKa = 3.23LSGAIEE118 pKa = 3.91ALRR121 pKa = 11.84EE122 pKa = 4.23GGLKK126 pKa = 10.14RR127 pKa = 11.84LRR129 pKa = 11.84EE130 pKa = 4.6DD131 pKa = 3.44CPEE134 pKa = 4.06VYY136 pKa = 10.19VKK138 pKa = 8.17YY139 pKa = 10.48HH140 pKa = 6.74RR141 pKa = 11.84GLHH144 pKa = 4.49QLYY147 pKa = 9.97NAEE150 pKa = 4.21LPSRR154 pKa = 11.84PRR156 pKa = 11.84APDD159 pKa = 3.08VHH161 pKa = 8.3LLFGPPGCGKK171 pKa = 8.28TRR173 pKa = 11.84FFYY176 pKa = 10.84DD177 pKa = 3.77NYY179 pKa = 10.13PNSPSLAVTDD189 pKa = 4.66GFWFDD194 pKa = 4.0GYY196 pKa = 10.97SGEE199 pKa = 4.75DD200 pKa = 3.0SVLIDD205 pKa = 5.2DD206 pKa = 4.74YY207 pKa = 11.67DD208 pKa = 4.06GKK210 pKa = 10.92ASRR213 pKa = 11.84WTLSNLLRR221 pKa = 11.84ILDD224 pKa = 3.96RR225 pKa = 11.84YY226 pKa = 9.88SIQVPIKK233 pKa = 10.64GGFTPWIPTRR243 pKa = 11.84IFITSNLHH251 pKa = 5.08PSNWYY256 pKa = 9.98DD257 pKa = 3.24YY258 pKa = 9.85SQRR261 pKa = 11.84QQRR264 pKa = 11.84YY265 pKa = 5.75QALSRR270 pKa = 11.84RR271 pKa = 11.84FTRR274 pKa = 11.84VTWWSSLQDD283 pKa = 4.06SEE285 pKa = 5.92DD286 pKa = 3.67LTPDD290 pKa = 3.26SPRR293 pKa = 11.84WEE295 pKa = 4.51HH296 pKa = 6.29FWAGPEE302 pKa = 4.06SAQRR306 pKa = 11.84SLDD309 pKa = 3.35IDD311 pKa = 3.58SGRR314 pKa = 11.84LISRR318 pKa = 11.84PPDD321 pKa = 3.35SEE323 pKa = 4.42FSFF326 pKa = 4.39
MM1 pKa = 7.57SRR3 pKa = 11.84GRR5 pKa = 11.84NWLFTEE11 pKa = 4.47NNPTLNPNDD20 pKa = 3.66FLEE23 pKa = 4.52SAKK26 pKa = 10.71LWDD29 pKa = 3.54HH30 pKa = 5.18VRR32 pKa = 11.84YY33 pKa = 10.0IIFQQEE39 pKa = 3.93TGEE42 pKa = 4.75GGTSHH47 pKa = 5.67YY48 pKa = 10.16QGYY51 pKa = 9.47VEE53 pKa = 4.96FSQPIRR59 pKa = 11.84FAKK62 pKa = 8.92VTGFLPRR69 pKa = 11.84AHH71 pKa = 6.55WEE73 pKa = 3.86MRR75 pKa = 11.84RR76 pKa = 11.84GTSAQCIEE84 pKa = 4.5YY85 pKa = 10.29CSKK88 pKa = 11.12LDD90 pKa = 3.65TRR92 pKa = 11.84VDD94 pKa = 4.17GPWTYY99 pKa = 11.41GDD101 pKa = 4.52PGFSSQGSRR110 pKa = 11.84SDD112 pKa = 3.23LSGAIEE118 pKa = 3.91ALRR121 pKa = 11.84EE122 pKa = 4.23GGLKK126 pKa = 10.14RR127 pKa = 11.84LRR129 pKa = 11.84EE130 pKa = 4.6DD131 pKa = 3.44CPEE134 pKa = 4.06VYY136 pKa = 10.19VKK138 pKa = 8.17YY139 pKa = 10.48HH140 pKa = 6.74RR141 pKa = 11.84GLHH144 pKa = 4.49QLYY147 pKa = 9.97NAEE150 pKa = 4.21LPSRR154 pKa = 11.84PRR156 pKa = 11.84APDD159 pKa = 3.08VHH161 pKa = 8.3LLFGPPGCGKK171 pKa = 8.28TRR173 pKa = 11.84FFYY176 pKa = 10.84DD177 pKa = 3.77NYY179 pKa = 10.13PNSPSLAVTDD189 pKa = 4.66GFWFDD194 pKa = 4.0GYY196 pKa = 10.97SGEE199 pKa = 4.75DD200 pKa = 3.0SVLIDD205 pKa = 5.2DD206 pKa = 4.74YY207 pKa = 11.67DD208 pKa = 4.06GKK210 pKa = 10.92ASRR213 pKa = 11.84WTLSNLLRR221 pKa = 11.84ILDD224 pKa = 3.96RR225 pKa = 11.84YY226 pKa = 9.88SIQVPIKK233 pKa = 10.64GGFTPWIPTRR243 pKa = 11.84IFITSNLHH251 pKa = 5.08PSNWYY256 pKa = 9.98DD257 pKa = 3.24YY258 pKa = 9.85SQRR261 pKa = 11.84QQRR264 pKa = 11.84YY265 pKa = 5.75QALSRR270 pKa = 11.84RR271 pKa = 11.84FTRR274 pKa = 11.84VTWWSSLQDD283 pKa = 4.06SEE285 pKa = 5.92DD286 pKa = 3.67LTPDD290 pKa = 3.26SPRR293 pKa = 11.84WEE295 pKa = 4.51HH296 pKa = 6.29FWAGPEE302 pKa = 4.06SAQRR306 pKa = 11.84SLDD309 pKa = 3.35IDD311 pKa = 3.58SGRR314 pKa = 11.84LISRR318 pKa = 11.84PPDD321 pKa = 3.35SEE323 pKa = 4.42FSFF326 pKa = 4.39
Molecular weight: 37.69 kDa
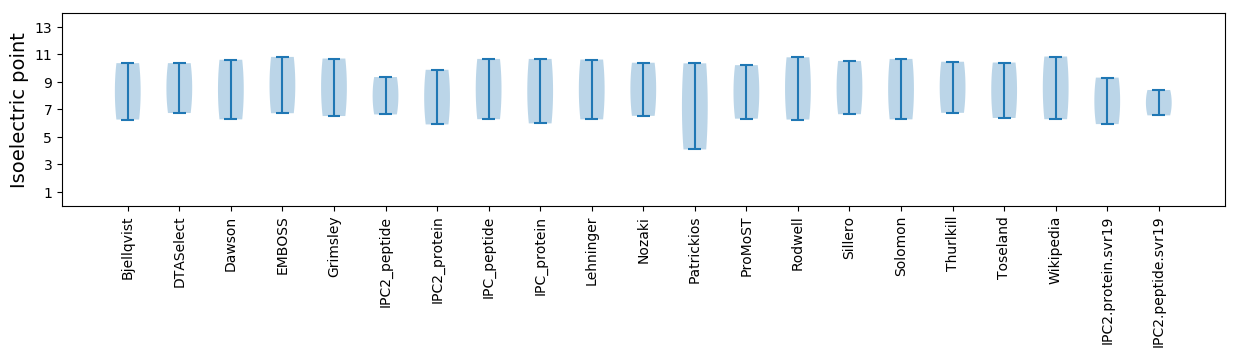
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPE8|A0A346BPE8_9VIRU Putative capsid protein OS=False black widow spider associated circular virus 1 OX=2293279 PE=4 SV=1
MM1 pKa = 6.97VRR3 pKa = 11.84YY4 pKa = 9.24SRR6 pKa = 11.84KK7 pKa = 9.06RR8 pKa = 11.84SFGRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84TFRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84NGLRR23 pKa = 11.84FKK25 pKa = 10.59RR26 pKa = 11.84RR27 pKa = 11.84VYY29 pKa = 10.2RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84TFRR35 pKa = 11.84GRR37 pKa = 11.84ARR39 pKa = 11.84GMQRR43 pKa = 11.84IQRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84YY49 pKa = 8.41PVNNPFGDD57 pKa = 3.72KK58 pKa = 10.43IFVKK62 pKa = 10.0FRR64 pKa = 11.84CTTGGNLQAPSVAAPSAGQTWNLNDD89 pKa = 4.28LGTAVGNGGAPGLEE103 pKa = 4.25EE104 pKa = 3.84YY105 pKa = 11.06SLLFKK110 pKa = 10.58RR111 pKa = 11.84YY112 pKa = 9.01RR113 pKa = 11.84ITGAKK118 pKa = 9.62VKK120 pKa = 9.04VTIYY124 pKa = 10.97NEE126 pKa = 3.82VSSDD130 pKa = 3.68VFNVDD135 pKa = 3.73PAWGFITASSDD146 pKa = 3.66AIDD149 pKa = 3.45MSDD152 pKa = 3.31YY153 pKa = 10.82HH154 pKa = 8.75LSDD157 pKa = 3.24IQHH160 pKa = 6.07LRR162 pKa = 11.84WVKK165 pKa = 10.15YY166 pKa = 10.28KK167 pKa = 10.36PLNNWTSGARR177 pKa = 11.84ATTISAYY184 pKa = 10.22FSCLKK189 pKa = 10.77VFGADD194 pKa = 2.97RR195 pKa = 11.84TIPDD199 pKa = 4.39DD200 pKa = 4.37LDD202 pKa = 3.58YY203 pKa = 11.04TATTNSAGTPFYY215 pKa = 8.67NTPSITAGVGWGVVKK230 pKa = 10.85ANASNWGASEE240 pKa = 5.29SIGQYY245 pKa = 7.44TQTVTYY251 pKa = 9.7YY252 pKa = 10.73AQLWDD257 pKa = 4.09PEE259 pKa = 4.37LGQTNVV265 pKa = 2.94
MM1 pKa = 6.97VRR3 pKa = 11.84YY4 pKa = 9.24SRR6 pKa = 11.84KK7 pKa = 9.06RR8 pKa = 11.84SFGRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84TFRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84NGLRR23 pKa = 11.84FKK25 pKa = 10.59RR26 pKa = 11.84RR27 pKa = 11.84VYY29 pKa = 10.2RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84TFRR35 pKa = 11.84GRR37 pKa = 11.84ARR39 pKa = 11.84GMQRR43 pKa = 11.84IQRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84YY49 pKa = 8.41PVNNPFGDD57 pKa = 3.72KK58 pKa = 10.43IFVKK62 pKa = 10.0FRR64 pKa = 11.84CTTGGNLQAPSVAAPSAGQTWNLNDD89 pKa = 4.28LGTAVGNGGAPGLEE103 pKa = 4.25EE104 pKa = 3.84YY105 pKa = 11.06SLLFKK110 pKa = 10.58RR111 pKa = 11.84YY112 pKa = 9.01RR113 pKa = 11.84ITGAKK118 pKa = 9.62VKK120 pKa = 9.04VTIYY124 pKa = 10.97NEE126 pKa = 3.82VSSDD130 pKa = 3.68VFNVDD135 pKa = 3.73PAWGFITASSDD146 pKa = 3.66AIDD149 pKa = 3.45MSDD152 pKa = 3.31YY153 pKa = 10.82HH154 pKa = 8.75LSDD157 pKa = 3.24IQHH160 pKa = 6.07LRR162 pKa = 11.84WVKK165 pKa = 10.15YY166 pKa = 10.28KK167 pKa = 10.36PLNNWTSGARR177 pKa = 11.84ATTISAYY184 pKa = 10.22FSCLKK189 pKa = 10.77VFGADD194 pKa = 2.97RR195 pKa = 11.84TIPDD199 pKa = 4.39DD200 pKa = 4.37LDD202 pKa = 3.58YY203 pKa = 11.04TATTNSAGTPFYY215 pKa = 8.67NTPSITAGVGWGVVKK230 pKa = 10.85ANASNWGASEE240 pKa = 5.29SIGQYY245 pKa = 7.44TQTVTYY251 pKa = 9.7YY252 pKa = 10.73AQLWDD257 pKa = 4.09PEE259 pKa = 4.37LGQTNVV265 pKa = 2.94
Molecular weight: 29.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
591 |
265 |
326 |
295.5 |
33.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.922 ± 1.662 | 1.015 ± 0.182 |
6.091 ± 0.828 | 3.723 ± 1.282 |
5.245 ± 0.237 | 8.46 ± 0.416 |
1.692 ± 0.654 | 4.23 ± 0.055 |
3.215 ± 0.654 | 6.937 ± 1.155 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
0.846 ± 0.2 | 4.569 ± 1.026 |
5.753 ± 1.119 | 3.892 ± 0.346 |
9.645 ± 0.643 | 8.799 ± 1.137 |
6.768 ± 1.334 | 4.907 ± 1.316 |
3.215 ± 0.4 | 5.076 ± 0.144 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
