
Brenneria alni
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Pectobacteriaceae; Brenneria
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
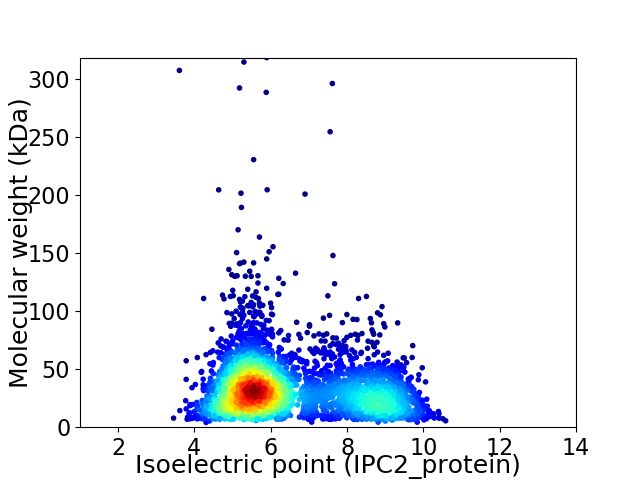
Virtual 2D-PAGE plot for 3626 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
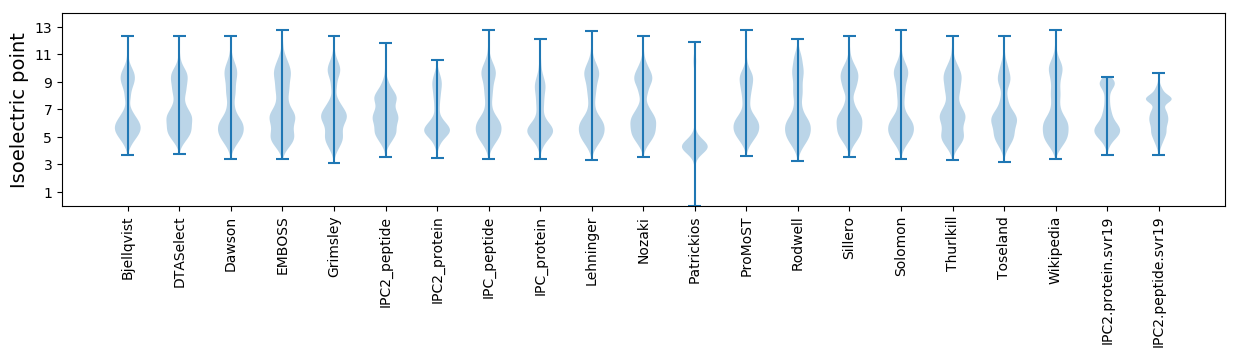
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A421DSC5|A0A421DSC5_9GAMM TPR_REGION domain-containing protein OS=Brenneria alni OX=71656 GN=BIY29_03140 PE=4 SV=1
MM1 pKa = 7.54GFSQAISGLNAASSNLDD18 pKa = 3.36VIGNNIANSEE28 pKa = 4.32TTGFKK33 pKa = 10.31AASVSFADD41 pKa = 4.24IFASSQVGLGVKK53 pKa = 9.33VASVSQDD60 pKa = 3.17FGDD63 pKa = 3.92GTVEE67 pKa = 3.99STDD70 pKa = 3.22RR71 pKa = 11.84GLDD74 pKa = 3.28VAIAGTGFFRR84 pKa = 11.84MTDD87 pKa = 3.49EE88 pKa = 4.58NGSVYY93 pKa = 10.45YY94 pKa = 10.51SRR96 pKa = 11.84NGEE99 pKa = 3.93LSLDD103 pKa = 3.69SEE105 pKa = 4.66RR106 pKa = 11.84NIVNSSGYY114 pKa = 10.48YY115 pKa = 7.39LTGYY119 pKa = 9.76AATGTPPTINTGAEE133 pKa = 3.77PTAINIPSGDD143 pKa = 3.19IAANATNSASMVLNLNSSADD163 pKa = 3.93TIPAGTTFDD172 pKa = 3.48ATDD175 pKa = 3.32ASTYY179 pKa = 10.35SYY181 pKa = 11.4SRR183 pKa = 11.84TLTTYY188 pKa = 10.65DD189 pKa = 3.47SQGNTHH195 pKa = 6.11NVNLYY200 pKa = 9.21FAKK203 pKa = 10.4TNDD206 pKa = 3.64NTWNVYY212 pKa = 10.07GVDD215 pKa = 3.74SSDD218 pKa = 3.9AAAEE222 pKa = 3.98PQTLGTLTFDD232 pKa = 3.65GSGMLTGTTEE242 pKa = 4.73LNVATNGINGSDD254 pKa = 3.96DD255 pKa = 3.24NTFAISFTGTTQQSASASTISSSTQDD281 pKa = 2.85GYY283 pKa = 10.98MAGSLSSYY291 pKa = 9.23TINSDD296 pKa = 3.27GTITGTYY303 pKa = 10.7SNGQTQLLGQIVMASFANPQGLKK326 pKa = 10.75AEE328 pKa = 4.58GDD330 pKa = 4.19NVWSSTASSGAAIVGTAGVGVFGSLTSGALEE361 pKa = 4.07SSNVDD366 pKa = 3.65LSAEE370 pKa = 4.11LVNMIVAQRR379 pKa = 11.84NYY381 pKa = 10.23QSNAQTIKK389 pKa = 9.47TQDD392 pKa = 3.73AILNTLVNLRR402 pKa = 4.14
MM1 pKa = 7.54GFSQAISGLNAASSNLDD18 pKa = 3.36VIGNNIANSEE28 pKa = 4.32TTGFKK33 pKa = 10.31AASVSFADD41 pKa = 4.24IFASSQVGLGVKK53 pKa = 9.33VASVSQDD60 pKa = 3.17FGDD63 pKa = 3.92GTVEE67 pKa = 3.99STDD70 pKa = 3.22RR71 pKa = 11.84GLDD74 pKa = 3.28VAIAGTGFFRR84 pKa = 11.84MTDD87 pKa = 3.49EE88 pKa = 4.58NGSVYY93 pKa = 10.45YY94 pKa = 10.51SRR96 pKa = 11.84NGEE99 pKa = 3.93LSLDD103 pKa = 3.69SEE105 pKa = 4.66RR106 pKa = 11.84NIVNSSGYY114 pKa = 10.48YY115 pKa = 7.39LTGYY119 pKa = 9.76AATGTPPTINTGAEE133 pKa = 3.77PTAINIPSGDD143 pKa = 3.19IAANATNSASMVLNLNSSADD163 pKa = 3.93TIPAGTTFDD172 pKa = 3.48ATDD175 pKa = 3.32ASTYY179 pKa = 10.35SYY181 pKa = 11.4SRR183 pKa = 11.84TLTTYY188 pKa = 10.65DD189 pKa = 3.47SQGNTHH195 pKa = 6.11NVNLYY200 pKa = 9.21FAKK203 pKa = 10.4TNDD206 pKa = 3.64NTWNVYY212 pKa = 10.07GVDD215 pKa = 3.74SSDD218 pKa = 3.9AAAEE222 pKa = 3.98PQTLGTLTFDD232 pKa = 3.65GSGMLTGTTEE242 pKa = 4.73LNVATNGINGSDD254 pKa = 3.96DD255 pKa = 3.24NTFAISFTGTTQQSASASTISSSTQDD281 pKa = 2.85GYY283 pKa = 10.98MAGSLSSYY291 pKa = 9.23TINSDD296 pKa = 3.27GTITGTYY303 pKa = 10.7SNGQTQLLGQIVMASFANPQGLKK326 pKa = 10.75AEE328 pKa = 4.58GDD330 pKa = 4.19NVWSSTASSGAAIVGTAGVGVFGSLTSGALEE361 pKa = 4.07SSNVDD366 pKa = 3.65LSAEE370 pKa = 4.11LVNMIVAQRR379 pKa = 11.84NYY381 pKa = 10.23QSNAQTIKK389 pKa = 9.47TQDD392 pKa = 3.73AILNTLVNLRR402 pKa = 4.14
Molecular weight: 41.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A421DQ17|A0A421DQ17_9GAMM EscR/YscR/HrcR family type III secretion system export apparatus protein OS=Brenneria alni OX=71656 GN=BIY29_07325 PE=3 SV=1
MM1 pKa = 7.06LTIAAHH7 pKa = 6.22CWLCRR12 pKa = 11.84LPLYY16 pKa = 10.28HH17 pKa = 6.84SRR19 pKa = 11.84HH20 pKa = 6.4GICSFCQRR28 pKa = 11.84HH29 pKa = 5.59LPQRR33 pKa = 11.84PLCCPRR39 pKa = 11.84CGLLSGDD46 pKa = 3.98PTRR49 pKa = 11.84QCGRR53 pKa = 11.84CLKK56 pKa = 10.48NPPPWQSITFVSDD69 pKa = 2.96YY70 pKa = 11.02VPPLNTLMKK79 pKa = 10.19QFKK82 pKa = 10.55FNGKK86 pKa = 7.55TEE88 pKa = 4.23LAPVLARR95 pKa = 11.84QILLRR100 pKa = 11.84WLTTYY105 pKa = 10.64RR106 pKa = 11.84EE107 pKa = 4.39HH108 pKa = 7.34ALSAPPHH115 pKa = 6.7LFRR118 pKa = 11.84PDD120 pKa = 3.32QLFTVPLHH128 pKa = 6.05QRR130 pKa = 11.84RR131 pKa = 11.84HH132 pKa = 4.09WSRR135 pKa = 11.84GFNQTEE141 pKa = 3.84LLARR145 pKa = 11.84CLTRR149 pKa = 11.84WLRR152 pKa = 11.84CDD154 pKa = 3.5YY155 pKa = 10.45QPRR158 pKa = 11.84AIARR162 pKa = 11.84IRR164 pKa = 11.84QTRR167 pKa = 11.84IQQKK171 pKa = 9.48LSASARR177 pKa = 11.84RR178 pKa = 11.84KK179 pKa = 8.6NLRR182 pKa = 11.84GAFSCNTALTGKK194 pKa = 9.85QVVLLDD200 pKa = 3.96DD201 pKa = 4.01VVTTGSTAAEE211 pKa = 3.99ISRR214 pKa = 11.84VLLAQGAAGVQVWCLCRR231 pKa = 11.84TLL233 pKa = 4.39
MM1 pKa = 7.06LTIAAHH7 pKa = 6.22CWLCRR12 pKa = 11.84LPLYY16 pKa = 10.28HH17 pKa = 6.84SRR19 pKa = 11.84HH20 pKa = 6.4GICSFCQRR28 pKa = 11.84HH29 pKa = 5.59LPQRR33 pKa = 11.84PLCCPRR39 pKa = 11.84CGLLSGDD46 pKa = 3.98PTRR49 pKa = 11.84QCGRR53 pKa = 11.84CLKK56 pKa = 10.48NPPPWQSITFVSDD69 pKa = 2.96YY70 pKa = 11.02VPPLNTLMKK79 pKa = 10.19QFKK82 pKa = 10.55FNGKK86 pKa = 7.55TEE88 pKa = 4.23LAPVLARR95 pKa = 11.84QILLRR100 pKa = 11.84WLTTYY105 pKa = 10.64RR106 pKa = 11.84EE107 pKa = 4.39HH108 pKa = 7.34ALSAPPHH115 pKa = 6.7LFRR118 pKa = 11.84PDD120 pKa = 3.32QLFTVPLHH128 pKa = 6.05QRR130 pKa = 11.84RR131 pKa = 11.84HH132 pKa = 4.09WSRR135 pKa = 11.84GFNQTEE141 pKa = 3.84LLARR145 pKa = 11.84CLTRR149 pKa = 11.84WLRR152 pKa = 11.84CDD154 pKa = 3.5YY155 pKa = 10.45QPRR158 pKa = 11.84AIARR162 pKa = 11.84IRR164 pKa = 11.84QTRR167 pKa = 11.84IQQKK171 pKa = 9.48LSASARR177 pKa = 11.84RR178 pKa = 11.84KK179 pKa = 8.6NLRR182 pKa = 11.84GAFSCNTALTGKK194 pKa = 9.85QVVLLDD200 pKa = 3.96DD201 pKa = 4.01VVTTGSTAAEE211 pKa = 3.99ISRR214 pKa = 11.84VLLAQGAAGVQVWCLCRR231 pKa = 11.84TLL233 pKa = 4.39
Molecular weight: 26.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1149954 |
38 |
3036 |
317.1 |
35.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.555 ± 0.049 | 1.057 ± 0.016 |
5.32 ± 0.032 | 5.607 ± 0.039 |
3.69 ± 0.029 | 7.263 ± 0.039 |
2.264 ± 0.021 | 6.024 ± 0.038 |
4.093 ± 0.034 | 11.029 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.594 ± 0.021 | 3.775 ± 0.028 |
4.452 ± 0.028 | 4.831 ± 0.037 |
5.933 ± 0.04 | 6.163 ± 0.035 |
5.271 ± 0.032 | 6.868 ± 0.033 |
1.366 ± 0.018 | 2.846 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
